ERCC1 expression in patients with colorectal cancer: a pilot study
Abstract
Aim: Excision repair cross complementation group 1 (ERCC1) has a key role in enhanced DNA damage repair caused by oxaliplatin-based therapy and may lead to resistance of these platinum drugs in colorectal cancer (CRC) patients. Hence, the present preliminary study aimed to explore the role of ERCC1 C/T polymorphism at codon 118 as well as its immunoreactivity in patients with primary CRC. Methods: ERCC1 polymorphism was studied using PCR-RFLP and ERCC1 protein expression was examined by immunohistochemistry in 50 CRC patients. Results: ERCC1 codon 118 C/T polymorphism analysis reported the predominance of C/T (52%) genotype as compared to C/C (38%) and T/T (10%) genotypes. Furthermore, 72% of patients showed positive ERCC1 protein expression. Significant correlation was not observed between clinicopathological parameters and ERCC1 polymorphism, while ERCC1 protein expression significantly correlated only with tumor site (colon vs. rectum) (P = 0.046). Further, the present study failed to demonstrate the role of ERCC1 C118T polymorphism or protein expression as useful prognostic markers in CRC patients. Conclusion: ERCC1-positive protein expression may be a useful marker for rectal cancer patients. However, further evaluation in a larger set of CRC patients is required to better understand the role of ERCC1.
Keywords
Introduction
The antimetabolite, 5-Flourouracil (5-FU), was introduced in 1957 by Heidelberger et al.[1] and today it is the cornerstone of chemotherapeutic regimens in treatment of colorectal cancer (CRC). However, the overall response rate is only 10-15% for advanced CRC when treated with 5-FU alone.[2] In recent years, the outcome of patients with CRC has been improved significantly because of the use of oxaliplatin-based combination therapy with 5-FU (FOLFOX). Oxaliplatin, in combination with leucovorin and 5-FU, is quite effective in the treatment of CRC, in both the adjuvant and metastatic settings.[3]
Oxaliplatin is a cytotoxic platinum compound which exerts its effects through development of DNA adducts.[4] DNA repair, especially nucleotide excision repair (NER) pathway, plays an important role in platinum-based chemotherapeutic efficacy by repairing drug-produced DNA damage. Excision repair cross complementation group 1 (ERCC1) is a chief component of the NER pathway[5,6] and a highly conserved protein, essential for elimination of DNA adducts caused by the platinum compound. It plays a major role in better repair and tolerance of DNA damage, leading to resistance of platinum drugs.[7,8]
A common C/T single nucleotide polymorphism (SNP) at codon 118 of ERCC1 has been identified. This SNP may contribute to inter-individual variability in DNA repair capability and has been documented as a predictor for outcome in CRC patients who have been treated with platinum-based chemotherapy.[9] This polymorphism results in the same amino acid, asparagine, but a tendency towards elevated ERCC1 mRNA and protein levels observed as the number of T alleles increases. Moreover, clinical and preclinical studies indicate that overexpression of ERCC1 protein is associated with resistance to platinum-based chemotherapy in various types of cancers.[10,11]
Therefore, the present study aimed to evaluate the prevalance of ERCC1 C118T SNP polymorphism and the expression of ERCC1 protein in patients with primary CRC and further to evaluate their role in the clinical outcome of CRC patients.
Methods
Patients
A total of 50 CRC patients who underwent surgical resection at the Gujarat Cancer and Research Institute, Ahmedabad, between 2013 and 2014 were included in this study. Inclusion criterion was an untreated CRC patient with histopathologically confirmed adenocarcinoma without any prior history of anticancer treatment. Patients were followed for a minimum of 15 months or until death within that period.
Sample collection
The study was approved by Institutional Scientific and Ethical Committees and informed consent was required from all patients prior to sample collection. To examine ERCC1 polymorphism, colorectal tumor was collected from the histopathology department immediately after surgery. The tumor portion was selected by the pathologist, snap frozen in liquid nitrogen, and immediately stored at -80 ℃ until analysis. For the study of ERCC1 protein expression, paraffin embedded tumor tissue blocks were retrieved from histopathology department.
DNA extraction and ERCC1 polymorphism study by PCR-RFLP
DNA extraction from tumor was performed by Phenol: Chloroform method. PCR of the SNP C/T at codon 118 of ERCC1 gene was performed using QIAGEN Taq PCR kit and following primers sequences: forward primer: 5’-GCAGAGCTCACCTGAGGAAC-3’; reverse primer: 5’-GAGGTGCAAGAAGAGGTGGA -3’ (Sigma-Aldrich). After initial denaturation at 94 °C for 3 min, the following PCR protocol was performed for 35 cycles: Denaturation at 95 °C for 1 min, annealing at 55.7 °C for 45 s, and extension at 72 °C extensions for 1 min. Digestion of these PCR products was performed with BsrD1 restriction enzyme (New England BioLabs, USA) at 65 °C for 4 h and separated on 2% agarose gel.
ERCC1 protein expression by immunohistochemistry
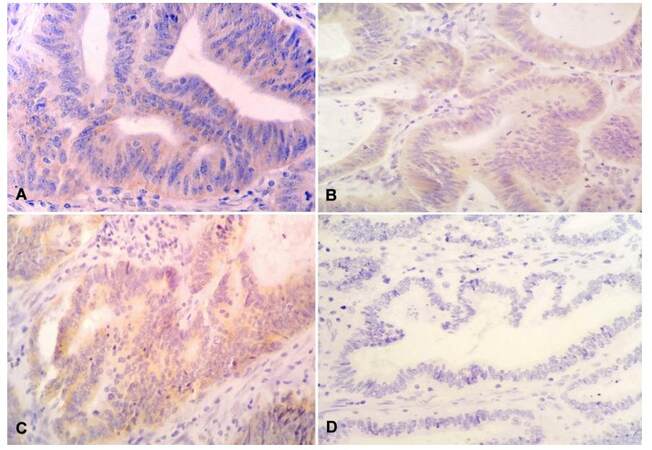
Immunohistochemistry for ERCC1 was performed on 4 μm thick formalin-fixed, paraffin embedded tissue sections. FFPE tumor tissue sections were stained using mouse- and rabbit- specific HRP/DAB (ABC) Detection IHC kit according to manufacturer’s protocol. The primary antibody used was mouse monoclonal anti-ERCC1 (clone 4F9: Dako, USA, dilution: 1:50). Sections known to exhibit high expression of protein were used as positive controls, while negative controls were obtained by omission of primary antibody. A semiquantitative score was used starting from negative (no staining or, 10% of cells stained) to 3+ (1+ staining for 11-30% of cells: weak, 2+ staining for 31-50% of cells: moderate, and 3+ staining for > 50% of cells: intense).
Statistical analysis
The statistical data analysis was performed by Statistical Package for the Social Sciences software version 17. Two tailed chi (χ2) test was used to determine the association between clinicopathological variables with ERCC1 polymorphism and ERCC1 protein expression. Correlation between two parameters was calculated according to Spearman’s correlation coefficient (r) method. Relapse-free survival (RFS) and overall survival (OS) was calculated using Log rank test. P value ≤ 0.05 was considered significant.
Results
Patient characteristics
Patient and tumor characteristics of the 50 patients are outlined in Table 1. The median age was 55 years. There was a considerable proportion (54%) of elderly and dominance of male patients (60%). The incidence of patients having colon cancer was higher (56%) as compared to rectal cancer (44%). Higher occurrence of early stage patients (60%) was observed. Complete follow-up details were obtained in 80% (40/50) and were included for OS analysis. Amongst these 40, 4 died due to disease and hence were not included for the RFS analysis. Therefore, 36/40 CRC patients were considered for RFS analysis from which 2 patients developed recurrence [Table 1].
Patient and tumor characteristics (n = 50)
| Characteristics | n (%) |
|---|---|
| Age (year) | |
| < 55 | 23 (46) |
| ≥ 55 | 27 (54) |
| Gender | |
| Female | 20 (40) |
| Male | 30 (60) |
| Habit | |
| No | 29 (58) |
| Yes | 21 (42) |
| Tumor site | |
| Colon | 28 (56) |
| Rectum | 22 (44) |
| Tumor size | |
| T2 | 5 (10) |
| T3 | 43 (86) |
| T4 | 2 (4) |
| TNM stage | |
| Early stage (I + II) | 30 (60) |
| Advanced stage (III + IV) | 20 (40) |
| Dukes’ stage | |
| B | 30 (60) |
| C | 20 (40) |
| Histological type | |
| Adenocarcinoma | 35 (70) |
| Mucin adenocarcinoma | 14 (28) |
| Signet ring cell carcinoma | 1 (2) |
| Histological grade | |
| Well differentiated | 9 (18) |
| Moderately differentiated | 33 (66) |
| Poorly differentiated | 8 (16) |
| CEA (ng/mL) (n = 45) | |
| < 5.0 | 19 (42) |
| ≥ 5.0 | 26 (58) |
| Treatment | |
| Surgery alone | 9 (18) |
| Surgery + chemotherapy | 25 (50) |
| Surgery + chemotherapy + radiotherapy | 12 (24) |
| Surgery + radiotherapy | 4 (8) |
| Recurrence/metastasis (n = 36) | |
| Absent | 34 (94) |
| Present | 2 (6) |
| Disease outcome (n = 40) | |
| Alive | 35 (88) |
| Dead | 5 (12) |
Incidence of ERCC1 codon 118 C/T polymorphism
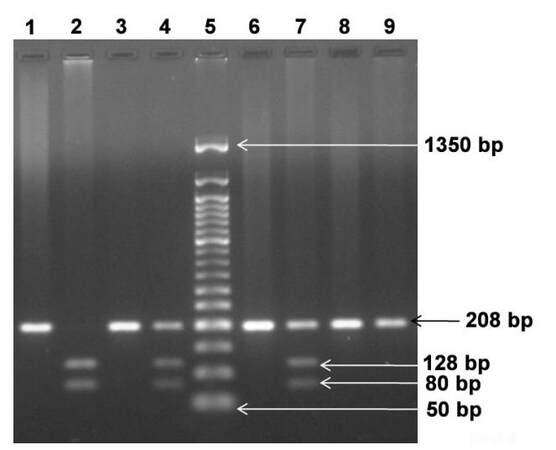
Three types of genotypes have been identified for ERCC1 C118T SNP: C/C genotype at 208 bp, C/T genotype at 208, 128 and 80 bp and T/T genotype at 128 and 80 bp [Figure 1].
Figure 1. Representative pattern of ERCC1 codon 118 genotypes separated on 2% agarose gel. Lane 1, 3, 6 and 8: undigested PCR products; Lane 2: T/T homozygous genotype at 128, 80 bp; Lane 4 and Lane 7: C/T heterozygous genotype at 208, 128 and 80 bp; Lane 9: C/C wild type genotype at 208 bp; Lane 5: 50 bp ladder. ERCC1: excision repair cross complementation group 1; PCR: polymerase chain reaction
Thirty-eight percent (n = 19) showed the C/C genotype, 52% (n = 26) showed the C/T genotype, and 10% (n = 5) showed the T/T genotype. Thus, prevalence of C/T heterozygous genotype was observed in this group of CRC patients. The distribution of ERCC1 codon 118 polymorphism genotypes was consistent with the Hardy-Weinberg equilibrium among patients (χ2 = 0.82, P = 0.36).
Incidence of ERCC1 protein expression
Expression of ERCC1 protein was localized in the cytoplasm of epithelial cells of colon and rectum [Figure 2]. ERCC1-positive protein expression was detected in 72% (n = 36) of patients whereas 28% (n = 14) showed ERCC1-negative protein expression.
Correlation of ERCC1 polymorphism and ERCC1 protein expression with clinicopathological parameters
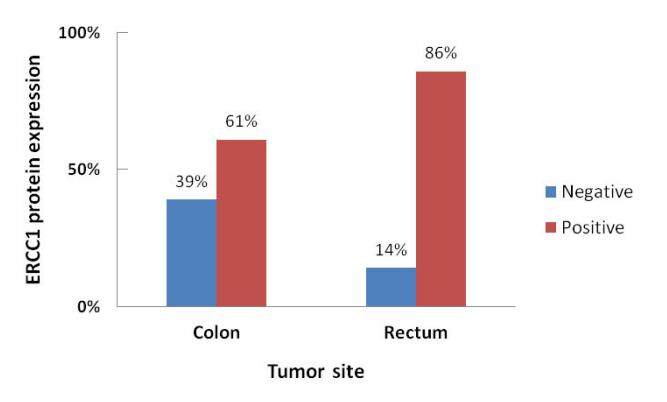
ERCC1 codon 118 C/T polymorphism was not significantly associated with any of the clinicopathological parameters. On the other hand, the incidence of ERCC1 protein immunoreactivity was significantly higher in patients having rectal (86%) than with colon cancer (61%) (χ2 = 4.020, r = +0.284, P = 0.046; Figure 3). ERCC1 protein expression was not significantly associated with the other parameters.
Correlation between ERCC1 polymorphism and ERCC1 protein expression
Predominance of ERCC1-positive protein expression was observed in all three sub-groups of patients having C/C (74% 14/19), C/T (69% 18/26), and T/T genotypes (80% 4/5). However, the difference was not statistically significant (P = 0.981).
Univariate survival analysis according to ERCC1 codon 118 C/T polymorphism and ERCC1 protein expression
When patients were stratified according to ERCC1-118 polymorphism, it was observed that both patients (100%) who had developed relapse had the C/T genotype, whereas amongst the 5 patients who died, 2 (40%) each had the C/C and C/T genotypes, and one (20%) had T/T. Further, when patients were stratified according to ERCC1 protein expression, of the 2 who developed recurrences, one (50%) was ERCC1-positive and another (50%) was ERCC1-negative. Among the patients who had died, 4 (75%) were ERCC1-positive and one (25%) was ERCC1 negative. However, the data were statistically non-significant.
Discussion
Oxaliplatin, a platinum-based chemotherapeutic drug, induces DNA damage by forming DNA adducts. DNA repair proteins involved in the NER pathway, such as ERCC1, play a key role in repair of this damage, thus leading to resistance to platinum-based therapy. Several clinical studies have demonstrated that both ERCC1 polymorphism and protein expression are associated with resistance to platinum-based chemotherapy and have the potential to be used as candidate biomarkers for CRC. Therefore, the present study explored the role of ERCC1 C118T SNP as well as ERCC1 immunoreactivity in CRC patients.
A predominance of the C/T genotype (52%) was observed as compared to C/C (38%) and T/T (10%) genotypes. In accordance with the current study, one CRC study reported a higher incidence of the C/T genotype, with 44% as compared to C/C (24%) and T/T (32%).[12] However, another reported that the frequencies of C/C, C/T, and T/T genotypes were 47.6%, 39.9% and 12.5%, respectively in Asian patients, very similar to previous reports in Caucasian populations.[4,13]
In the present study, it was found that there was positive ERCC1 protein expression in 72% of CRC patients. Likewise Wang et al.[14] reported 68.1% positive ERCC1 immunoreactivity in patients with gastric cancer. However, in 2 studies, ERCC1 positivity was observed in 45% and 55% of patients with colorectal and stage III disease, respectively.[4,15] This discrepancy may be due to several factors including antibody used, scoring technique, used and in preparation of the paraffin embedded tissue blocks.
To date, most studies have focused more on pathologic and molecular features, but less on clinical features with molecular variables. Therefore, in the present study, correlation of ERCC1 polymorphism and protein expression with clinicopathological parameters was evaluated. However, none of the clinicopathological parameters was significantly associated with ERCC1 gene polymorphism. Similarly, another study did not find statistically significant correlations between genotype distributions and gender, tumor location, tumor invasion, lymph node metastasis, tumor stage, or histology in CRC patients.[16] The present study showed no significant association between ERCC1 protein expression and clinicopathological parameters apart from tumor site. Still another study reported no significant differences in gender, tumor stage, nodal stage, histological differentiation, lympho-vascular invasion, neural invasion, or postoperative CEA levels between the ERCC1 positive and negative groups. However, ERCC1 positive expression was significantly associated with older age group patients (P = 0.031).[17] Also it has been shown in patients with nasopharyngeal carcinoma that the expression level of ERCC1 increased significantly with higher T stage and clinical stages (P < 0.05). Thus, at least in that malignancy, ERCC1 seemed to be a valid biological indicator to predict prognosis.[18] In the present study, ERCC1 positive protein expression was found to be significantly higher only in rectal cancer patients as compared to colon cancer patients (P = 0.046).
In one report the identical ERCC1 C/T polymorphism at codon 118 was found to influence the level of ERCC1 expression. This may be due to that, although both the AAC and AAT codons encode asparagine, the AAT codon usage is significantly reduced, thereby decreasing ERCC1 translation capability and protein level.[19] Therefore, in the present study we correlated ERCC1-118 polymorphism and ERCC1 protein expression. However, there was no significant correlation found between ERCC1 polymorphism and ERCC1 protein expression. Similarly, Qi et al.[20] in patients with gastric cancer and Takenaka et al.[21] in patients with non-small cell lung cancer also showed that ERCC1 genotypes were not correlated with ERCC1 protein expression. On the other hand, another study found increased ERCC1 protein levels in CRC patients with the C/T or T/T genotypes.[4]
Several studies have investigated the prognostic role of ERCC1-118 SNP and ERCC1 protein expression in CRC patients and other cancers types but the results obtained were controversial. In one report ERCC1 codon 118 C/C genotype was significantly associated with higher response rates, progression-free survival, and OS in metastatic CRC.[4] However, another supported the pharmacogenetic role of the ERCC1-118 C > T change and emphasized that the T allele was a marker of a better outcome in patients with CRC treated with OX-based schemes.[22,23] Thus, the relationship between ERCC1 codon 118 SNP and clinical outcome in patients with CRC remains controversial although one study reported significantly shorter progression-free (P < 0.01) and overall (P < 0.01) survival in patients having positive ERCC1 IHC staining in colorectal tumor tissues.[4] It was also observed that 5-year DFS and OS were significantly lower in combined therapy group with positive ERCC1 tumors than in the same group patients with negative ERCC1 tumors.[15] However, in present study, due to short follow-up period of 15 months, no correlation of RFS and OS was observed with ERCC1 C118T SNP and ERCC1 protein expression. We recognize that a limitation of this study is that SNP assessment was not confirmed by Sanger sequencing. Further studies will be necessary to validate our findings.
In summary, the current study reveals that ERCC1 protein expression was significantly higher in patients with rectal cancer as compared to patients with colon cancer, which may indicate biological and functional differences between the two subsets and may emerge as an important marker for patients with rectal cancer. However, further studies with larger sample sizes and longer follow-up period are necessary for a more definite conclusion.
Financial support and sponsorship
This work was financially supported by Gujarat Cancer Society (GCS)/Gujarat Cancer and Research Institute (GCRI) and Directorate of Medical Education and Research (DMER).
Conflicts of interest
There are no conflicts of interest.
Patient consent
Patient consent was acquired from all patients prior to sample collection.
Ethics approval
The study has been approved by Institutional Scientific and Ethical Committees.
REFERENCES
1. Heidelberger C, Chaudhuri NK, Danneberg P, Mooren D, Griesbach L, Duschinsky R, Schnitzer RJ, Pleven E, Scheiner J. Fluorinated pyrimidines, a new class of tumour-inhibitory compounds. Nature 1957;179:663-6.
2. Zhang N, Yin Y, Xu SJ, Chen WS. 5-Fluorouracil: mechanisms of resistance and reversal strategies. Molecules 2008;13:1551-69.
3. de Gramont A, Figer A, Seymour M, Homerin M, Hmissi A, Cassidy J, Boni C, Cortes-Funes H, Cervantes A, Freyer G, Papamichael D, Le Bail N, Louvet C, Hendler D, de Braud F, Wilson C, Morvan F, Bonetti A. Leucovorin and fluorouracil with or without oxaliplatin as first-line treatment in advanced colorectal cancer. J Clin Oncol 2000;18:2938-47.
4. Chang PM, Tzeng CH, Chen PM, Lin JK, Lin TC, Chen WS, Jiang JK, Wang HS, Wang WS. ERCC1codon 118 C→T polymorphism associated with ERCC1 expression and outcome of FOLFOX-4 treatment in Asian patients with metastatic colorectal carcinoma. Cancer Sci 2009;100:278-83.
5. Dabholkar M, Bradshaw L, Parker RJ, Gill I, Bostick-Bruton F, Muggia FM, Reed E. Cisplatin-DNA damage and repair in peripheral blood leukocytes in vivo and in vitro. Environ Health Perspect 1992;98:53-9.
6. De Silva IU, McHugh PJ, Clingen PH, Hartley JA. Defining the roles of nucleotide excision repair and recombination in the repair of DNA interstrand cross-links in mammalian cells. Mol Cell Biol 2000;20:7980-90.
7. Raymond E, Faivre S, Woynarowski JM, Chaney SG. Oxaliplatin: mechanism of action and antineoplastic activity. Semin Oncol 1998;25:4-12.
8. Altaha R, Liang X, Yu JJ, Reed E. Excision repair cross complementing-group 1: gene expression and platinum resistance. Int J Mol Med 2004;14:959-70.
9. Park DJ, Zhang W, Stoehlmacher J, Tsao-Wei D, Groshen S, Gil J, Yun J, Sones E, Mallik N, Lenz HJ. ERCC1 gene polymorphism as a predictor for clinical outcome in advanced colorectal cancer patients treated with platinum-based chemotherapy. Clin Adv Hematol Oncol 2003;1:162-6.
10. Kwon HC, Roh MS, Oh SY, Kim SH, Kim MC, Kim JS, Kim HJ. Prognostic value of expression of ERCC1, thymidylate synthase, and glutathione S-transferase P1 for 5-fluorouracil/oxaliplatin chemotherapy in advanced gastric cancer. Ann Oncol 2007;18:504-9.
11. Ferry KV, Hamilton TC, Johnson SW. Increased nucleotide excision repair in cisplatin-resistant ovarian cancer cells: role of ERCC1-XPF. Biochem Pharmacol 2000;60:1305-13.
12. Zaanan A, Dalban C, Emile JF, Blons H, Fléjou JF, Goumard C, Istanbullu M, Calmel C, Alhazmi K, Validire P, Louvet C. ERCC1, XRCC1 and GSTP1 single nucleotide polymorphisms and survival of patients with colon cancer receiving oxaliplatin-based adjuvant chemotherapy. J Cancer 2014;5:425-32.
13. Stoehlmacher J, Park DJ, Zhang W, Yang D, Groshen S, Zahedy S, Lenz HJ. A multivariate analysis of genomic polymorphisms: prediction of clinical outcome to 5-FU/oxaliplatin combination chemotherapy in refractory colorectal cancer. Br J Cancer 2004;91:344-54.
14. Wang AT, Sengerová B, Cattell E, Inagawa T, Hartley JM, Kiakos K, Burgess-Brown NA, Swift LP, Enzlin JH, Schofield CJ, Gileadi O, Hartley JA, McHugh PJ. Human SNM1A and XPF-ERCC1 collaborate to initiate DNA interstrand cross-link repair. Genes Dev 2011;25:1859-70.
15. Li Y, Liu Z, Liu H, Wang LE, Tan D, Ajani JA, Wei QY. ERCC1 and ERCC2 variants predict survival in gastric cancer patients. PLoS One 2013;8:e71994.
16. Huang MY, Wang JY, Huang ML, Chang HJ, Lin SR. Polymorphisms in XPD and ERCC1 associated with colorectal cancer outcome. Int J Mol Sci 2013;14:4121-34.
17. Kim CY, Seo SH, An MS, Kim KH, Bae KB, Hwang JW, Kim JH, Kim BM, Kang MS, Oh MK, Hong KH. ERCC1 as a predictive marker for FOLFOX chemotherapy in an adjuvant setting. Ann Coloproctol 2015;31:92-7.
18. Liang R, Lin Y, Liu ZH, Liao XL, Yuan CL, Liao SN, Li YQ. Correlation between ERCC1 expression and concurrent chemotherapy and radiotherapy in patients with locally advanced nasopharyngeal cancer. Genet Mol Res 2015;14:5804-11.
19. Yu JJ, Thornton K, Guo Y, Kotz H, Reed E. An ERCC1 splicing variant involving the 5′UTR of the mRNA may have a transcriptional modulatory function. Oncogene 2001;20:7694-8.
20. Qi YJ, Cui S, Yang YZ, Han JQ, Cai BJ, Sheng CF, Ma Y, Wuren T, Ge RL. Excision repair cross-complementation group 1 codon 118 polymorphism, micro ribonucleic acid and protein expression, clinical outcome of the advanced gastric cancer response to first-line FOLFOX-4 in Qinghai-Tibetan plateau population. J Cancer Res Ther 2013;9:410-5.
21. Takenaka T, Yano T, Kiyohara C, Miura N, Kouso H, Ohba T, Kometani T, Shoji F, Yoshino I, Maehara Y. Effects of excision repair cross-complementation group 1 (ERCC1) single nucleotide polymorphisms on the prognosis of non-small cell lung cancer patients. Lung Cancer 2010;67:101-7.
22. Viguier J, Boige V, Miquel C, Pocard M, Giraudeau B, Sabourin JC, Ducreux M, Sarasin A, Praz F. ERCC1 codon 118 polymorphism is a predictive factor for the tumor response to oxaliplatin/5-fluorouracil combination chemotherapy in patients with advanced colorectal cancer. Clin Cancer Res 2005;11:6212-7.
23. Martinez-Balibrea E, Abad A, Aranda E, Sastre J, Manzano JL, Díaz-Rubio E, Gómez-Espa-a A, Aparicio J, García T, Maestu I, Martínez-Cardús A, Ginés A, Guino E; Spanish Group for the Treatment of Digestive Tumours. Pharmacogenetic approach for capecitabine or 5-fluorouracil selection to be combined with oxaliplatin as first-line chemotherapy in advanced colorectal cancer. Eur J Cancer 2008;44:1229-37.
Cite This Article
Export citation file: BibTeX | RIS
OAE Style
Gajjar KK, Yadav DK, Kobawala TP, Trivedi TI, Vora HH, Ghosh NR. ERCC1 expression in patients with colorectal cancer: a pilot study. J Cancer Metastasis Treat 2016;2:471-6. http://dx.doi.org/10.20517/2394-4722.2016.52
AMA Style
Gajjar KK, Yadav DK, Kobawala TP, Trivedi TI, Vora HH, Ghosh NR. ERCC1 expression in patients with colorectal cancer: a pilot study. Journal of Cancer Metastasis and Treatment. 2016; 2: 471-6. http://dx.doi.org/10.20517/2394-4722.2016.52
Chicago/Turabian Style
Gajjar, Kinjal K., Deep Kumari Yadav, Toral P. Kobawala, Trupti I. Trivedi, Hemangini H. Vora, Nandita R. Ghosh. 2016. "ERCC1 expression in patients with colorectal cancer: a pilot study" Journal of Cancer Metastasis and Treatment. 2: 471-6. http://dx.doi.org/10.20517/2394-4722.2016.52
ACS Style
Gajjar, KK.; Yadav DK.; Kobawala TP.; Trivedi TI.; Vora HH.; Ghosh NR. ERCC1 expression in patients with colorectal cancer: a pilot study. J. Cancer. Metastasis. Treat. 2016, 2, 471-6. http://dx.doi.org/10.20517/2394-4722.2016.52
About This Article
Copyright
Data & Comments
Data
 Cite This Article 8 clicks
Cite This Article 8 clicks











Comments
Comments must be written in English. Spam, offensive content, impersonation, and private information will not be permitted. If any comment is reported and identified as inappropriate content by OAE staff, the comment will be removed without notice. If you have any queries or need any help, please contact us at support@oaepublish.com.