The role of long noncoding RNAs in cancer metastasis
Abstract
Signaling pathways are tightly controlled systems that regulate the appropriate timing of gene expression required for the differentiation of cells down a particular lineage essential for proper tissue development. Proliferation, apoptosis and metabolic pathways are just a few examples of the signaling pathways that require fine-tuning, so as to control the proper development of a particular tissue type or organ system. An estimated 70% of the genome is actively transcribed, only 2% of which codes for known protein-coding genes. Long noncoding RNAs (lncRNAs) in particular, are a large and diverse class of RNAs > 200 nucleotides in length, and not translated into protein. lncRNAs are essential transcriptional and post-transcriptional regulators that control the expression of genes in a spatial, temporal, and cell context-dependent manner. The aberrant expression of lncRNAs is therefore linked with a number of chronic diseases including cardiac dysfunction, diabetes, and cancer. In this review, we highlight the specific role lncRNAs have in promoting the metastatic cascade across a number of epithelial cancer models.
Keywords
Introduction
Metastasis is the leading cause of cancer-related deaths world-wide[1]. Understanding the biological processes that control the initiation and progression of metastasis is crucial in reducing tumor-related deaths associated with carcinomas[2,3]. Metastasis consists of the following phases: (1) escape of cells from the primary tumor and invasion into the surrounding mesenchyme[4]; (2) intravasation into adjacent vasculature and the lymphatic system[5]; (3) upregulation of cell survival mechanisms via resistance to apoptosis and anoikis[6]; (4) extravasation from the vasculature and subsequent infiltration into the parenchyma of a distant organ site[7]; and (5) the ability to undergo micro-metastatic colonization, and survival within a new tissue microenvironment[8,9]. The epithelial to mesenchymal transition (EMT) is a key developmental regulatory program describing the initiating processes of metastasis, and involves a linear series of events including tightly organized epithelial cells undergoing a loss of cellular polarity, and the ability for cells to survive under anchorage-independent conditions, both of which supports the propagation of migratory cells able to invade distant organ sites[10]. EMT essentially reactivates the embryonic morphogenesis and wound healing programs normally kept inactive within differentiated epithelial cells[11-13]. Therefore, investigating the series of cellular reprogramming events required for differentiated epithelial cells to acquire an invasive mesenchymal phenotype will aid in the development of therapeutics that specifically target metastatic cells.
While many zinc finger transcription factors (TFs) have been identified as regulators of EMT, including zinc-finger enhancer binding 1 (ZEB1), Snail, and Slug, little is known regarding the initiating steps that drive the transition of polar cells of an epithelial origin towards those with mesenchymal characteristics[14,15]. Furthermore, given invasive metastatic cells hone to various tissue sites depending upon the tissue of origin from which the primary tumor derives (i.e., the “seed and soil hypothesis”), one can hypothesize that ubiquitously expressed TFs such as Snail cannot be the sole contributor of a cell-context dependent regulatory process such as metastasis[16,17]. In fact, in a recent survey of the human genomic landscape, there is striking evidence that noncoding RNAs (ncRNAs) play an important and diverse role in regulating developmental transitions. Moreover, ncRNAs control the spatial and temporal tuning of cellular signaling pathways important for the proper execution of functional phenotypes such as enhanced cellular proliferation, migration, and/or survival[18-26]. Furthermore, in cancers that are dependent upon changes in the abundance and bioavailability of steroid hormones such as 17β-estradiol, ncRNAs have been identified to play a key role in the abrogating hormone-mediated metastasis[19,27-33].
Therefore, ncRNAs are considered important epigenetic regulators of the transcriptome that modulate context-specific processes involved in promoting a metastatic phenotype. One class of ncRNA includes microRNAs (miRNAs), which are short 22-nucleotide (nt) ncRNAs that undergo biochemical processing from a longer primary miRNA (pri-miRNA) transcript via a series of interactions with RNase-III type proteins that include DROSHA and DICER. miRNAs operate via a distinct mechanism of action that relies upon imperfect complementarity or Watson-Crick base-pairing between a miRNA and the 3’ untranslated region (3’ UTR) of a target messenger RNA (mRNA)[34]. miRNAs therefore serve as guides that recruit RNA binding proteins (RBPs) such as AGO2 to specific mRNA targets resulting in reduced gene expression either via translational inhibition or via RNA degradation[25,35-37].
Given this imperfect complementarity, miRNAs function as pleiotropic regulators of cell signaling pathways critically important in maintaining proper tissue development, as well as inhibiting the initiation and progression of tumorigenic cascades[38]. Given miRNAs operate by fine-tuning gene expression, and themselves function as either oncogenes or tumor suppressors when dysregulated, these ncRNAs subsequently present as potential targets for therapeutic development across a wide number of genetic disorders. miRNAs also modulate the expression of genes considered initiators of EMT, as well as mediators of downstream metastatic processes such as micro-metastatic colonization, anoikis, and interactions within the surrounding tumor microenvironment. For instance, miR-10b is a miRNA expressed at high levels in metastatic breast cancer samples, and the use of anti-miRNA oligonucleotides significantly reduces metastatic lesions in mouse models[39,40]. Additionally, miRNAs such as miR-148a regulate the levels of E-cadherin and subsequently the progression of EMT via the modulation of DNMT1 activity[41], while the miR-17 family of miRNAs controls metastatic phenotypes in lung cancer via dampening the expression of transforming growth factor (TGF)-β[42]. Comprehensive reviews of miRNA in cancer are discussed in greater detail elsewhere[43,44].
lncRNAs are a newly discovered class of ncRNA important in dampening stochastic gene expression by modulating the epigenetic landscape of the genome. lncRNAs are a divergent class of ncRNA molecule greater than 200 nt in length that lack protein-coding capacity, yet control a diverse array of biological processes via the recruitment of chromatin modifiers to specific genomic loci or by modulating post-transcriptional processes[45,46]. Currently there are over 118,000 high confidence lncRNA transcripts identified in Homo sapiens (http://www.lncipedia.org), many of which have no ascribed biological function. However, a number of studies have begun elucidating particular ncRNAs dysregulated across multiple cancer types[19,47-49]. The challenge in studying lncRNAs is their relatively low abundance and reduced conservation across species as compared to protein coding transcripts and other ncRNAs such as miRNAs[50]. This led many to believe that lncRNAs derive from leaky transcriptional processes and, therefore, have limited functionality in regulating cellular processes. However, there is considerable evidence that lncRNAs regulate the physiological pathways required for the initiation and maintenance of the metastatic process.
Broadly speaking, the diminished level of a lncRNA within a cell, results in the reduced bioavailability of a particular enzymatic substrate important in modulating chromatin structure as well as the transcriptional activity of neighboring protein coding genes. This occurs via a chaperone mechanism whereby a lncRNA brings into proximity RBPs, as well as components of the transcriptional machinery including RNA polII, to discrete genetic loci facilitating proper TF binding[35,45,51-58]. Therefore, the abundance of any particular lncRNA is important in providing the specificity necessary to promote certain phenotypic outcomes required during the metastatic cascade[59,60]. While investigators have identified specific roles for lncRNAs that control a number of cellular functions including differentiation, invasion, and metastasis, this review focuses on the role of lncRNAs within the metastatic process [Table 1].
lncRNAs associated with and tumorigenesis and EMT pathways
| lncRNA | Cancer type | Expression in cancer | References | Role in metastasis |
|---|---|---|---|---|
| MALAT1 | Lung | Upregulated | [190] | Suppression of E-cadherin[191] |
| Bladder | Upregulated | [192] | ||
| Breast | Upregulated | [193] | ||
| Pancreatic | Upregulated | [194] | ||
| MEG3 | Meningioma | Downregulated | [195] | Regulation of autophagy[196] and DNA repair[197] |
| Lung | Downregulated | [198] | ||
| Gastric | Downregulated | [199] | ||
| HOTAIR | Liver | Upregulated | [200] | Reprogramming of chromatin state[201,202] |
| Breast | Upregulated | [203] | ||
| Pancreatic | Upregulated | [204] | ||
| GAS5 | Liver | Downregulated | [205] | Controls invasion by control of miRNAs[205,206] |
| Gastric | Downregulated | [207] | ||
| Breast | Downregulated | [208] | ||
| H19 | Liver | Upregulated | [209] | Chromatin remodeling[210] and TGF-β regulation[211] |
| Pancreatic | Upregulated | [137,212] | ||
| Gastric | Upregulated | [213] | ||
| HULC | Liver | Upregulated | [147] | Regulates tumor microenvironment interactions[214] |
| Gastric | Upregulated | [148] | ||
| Breast | Upregulated | [92,146] | ||
| SPRY4-IT1 | Melanoma | Upregulated | [139] | Proliferation and invasion via regulation of EZH2[143] |
| Lung | Upregulated | [215] | ||
| SCC | Upregulated | [142] |
LncRNA nomenclature
lncRNAs are a heterogeneous class of ncRNA transcribed from a number of regions within the genome, and in varying orientations that flank neighboring protein coding genes, promoting a diverse combination of functional phenotypes. The nomenclature of lncRNAs is still controversial; however, a concerted effort has been made to group lncRNAs into functional categories based on the genomic localization of these transcripts, as well as the regulatory functions they confer [Figure 1]. For instance, promoter-associated lncRNAs (pa-lncRNAs) are transcribed in an antisense orientation from a shared promoter of a neighboring protein coding gene[61]. A majority of pa-lncRNAs operate in cis and recruit chaperone proteins that modulate the transcription of the neighboring protein coding gene, though this is not always the case[62,63]. For instance, a pa-lncRNA was found to be transcribed from the cyclin D1 promoter and is important in mediating the inhibitory activity of certain histone acetyltransferases[64].
Figure 1. lncRNAs derive from a number of genetic loci and associate with specific lncRNA function. (A) pa-lncRNAs originate from a bi-directional promoter from the sense strand of gene foci. These lncRNAs tend to operate in cis and regulate the neighboring protein coding gene; (B) ea-lncRNAs are similar to pa-lncRNAs yet are transcribed from enhancer regions within the genome; (C) NAT-lncRNAs are transcribed from the antisense strand and contain fully or partially complementary sequences to sense-strand transcripts, depending upon the surrounding genetic elements that regulate transcription of NATs; (D) gb-lncRNAs are transcribed in sense orientation, typically are one exon in length, and could share exons from protein coding transcripts; (E) lincRNAs are transcribed from genetic loci in either sense or antisense fashion and span regions considered transcriptionally active, coding or otherwise. Portions of this figure were adapted from Martens-Uzunova et al.[229], with permission
Enhancer-associated lncRNAs (ea-lncRNAs) are similar to pa-lncRNAs, yet they originate from active enhancer regions within the genome that promote cis-activation of transcription via DNA looping at the proximal promoters of nearby protein coding genes[65]. ea-lncRNAs are also released from the site of transcription, and modulate the activity of distal gene promoters in trans through the recruitment of co-activators such as, p300/cAMP response element-binding protein (CREBP), as well as, demethylases such as lysine-specific demethylase 1 (LSD1)[66,67]. As an example, Braveheart (Bvht) is a lncRNA transcribed from an enhancer region marked by H3K27Ac, associates with cardiac specific transcriptional enhancers, and when lost in mice perturbs the development of cardiomyocytes indicating Bvht is an important regulator of mammalian cardiac development[67,68].
Natural antisense transcripts (NATs) are considered full length RNA transcripts initiated on the antisense strand of a respective protein coding gene[69,70]. Given this type of lncRNA has high complementarity to the mRNA transcript deriving from the sense strand, the formation of localized RNA duplexes results in enhanced RNA stability through HuR binding, or degradation via activation of RNA interference (RNAi) pathways. HIF1A-AS2, for instance, is transcribed from the HIF1A locus and operates as a scaffold, recruiting chromatin remodeling complexes, as well as RBPs such as IGF2BP2 to distinct genetic loci[71]. HIF1A-AS2 is also important in regulating hypoxic responses in A549 lung cancer cells[72]. In fact, hypoxia induces HIF1A-AS2 expression, which in turn binds and represses HIF1α levels under hypoxic conditions presumably through a process of RNA-mediated decay.
Gene body associated lncRNAs (gba-lncRNAs) differ from NATs, and originate instead from the sense strand of a respective protein coding gene loci[73-75]. An example of a gba-lncRNA is the pseudogene transcribed from the CCAAT/enhancer binding protein alpha (C/EBPα) locus, termed ecCEBPA, which utilizes a separate open reading frame (ORF) and transcriptional start site (TSS) neighboring the C/EBPα gene locus[76]. ecCEBPA interacts with DNMT1, resulting in decreased methylation of the CEBPA gene. Mutagenesis studies further indicated that ecCEBPA contains hairpin structures that favor DNMT1 binding suggesting lncRNAs are important in modulating not only transcriptome wide DNA methylation, but are also present at sites of active transcription. Other gba-lncRNAs can operate as sponges for ncRNAs, thereby modulating the bioavailability of mRNA transcripts within a cell[73,74]. This competitive endogenous RNA (ceRNA) code or hypothesis is discussed in greater detail later in the review.
Finally, long intergenic noncoding RNAs (lincRNA) span extensive regions of the genome and are found within intronic regions of a coding gene, rather than as discrete genetic elements[71]. Examples include HOTAIR and MALAT1 [Figure 1]. One of the first lincRNAs discovered, X inactive specific transcript (XIST), produces an approximately 20 kilobase (kb) noncoding lincRNA and functions to silence the expression of genes derived from the inactive X chromosome (Xi) through recruitment of the polycomb repressive complex (PRC1/2)[77]. The precise mechanism by which XIST recruits PRC1/2 to the X-chromosome is still unclear, as X-inactivation requires an evenly distributed presence of PRC1/2 across the Xi so as to ensure the proper silencing of both coding and noncoding transcripts [Figure 2B]. Some have indicated that the silencing of Xi is accompanied by phosphorylation events on p53, indicating XIST cooperates with the p53 DNA-repair machinery during X-inactivation[78]. Given other ncRNAs such as miR-34 are known regulators of TP53 expression in cancer cell lines as well as during development[79], this raises the notion that lincRNAs cooperate with ncRNAs to carry out specific cellular programs. XIST also mediates epigenetic interactions between PRC1/2 and specific gene loci via interactions with chromatin modifiers such as SHARP, SAF-A, and LBR to initiate transcriptional silencing[77]. Taken together, it is crucial that these biochemistry-focused studies continue, such that, novel therapies can be developed to modulate a specific biological activity mediated by a particular lncRNA of interest.
Figure 2. lncRNAs regulate transcriptional and post-transcriptional processes important in modulating gene expression. (A) Depicts the number of interactions lncRNAs have with other ncRNAs to modulate the ceRNA network. Specifically, lncRNAs can interact with miRNA recruiting these small RNAs from cognate mRNA targets. miRNAs can also compete for lncRNA or mRNA target binding depending upon the respective transcript abundance. Finally, lncRNAs can alter the stability of mRNAs either by recruiting RBPs such as HuR, or by preventing miRNA-mediated mRNA degradation; (B) represents a number of interactions that modulates the chromatin-architecture, such as MALAT1 regulation of the PRC1 complex that can modulate the euchromatin state. Additionally, XIST can recruit PRC2 to chromatin sites that preclude RNAPII chromatin binding. Finally, lncRNA ROR sponge histone methyltransferases away from heterochromatic regions, promoting transcription and (C) depicts a special chromatin modulation termed "chromosomal looping" which brings seemingly distance chromosomal regions into proximity for transcriptional control under cis-regulatory interactions. Chromosomal looping also favors additional chromatin modifications to occur at specific genomic locations. Parts of figure are adapted from Long et al.[66], with permission
Lncrna mechanism of action
lncRNAs communicate with other ncRNAs via the “ceRNA code”
The ceRNA hypothesis, specifically the notion that RNA-RNA interactions operate in a complex regulatory pattern through competitive Watson-Crick base-pairing interactions, formed after the discovery that PTENP1, a particular ncRNA with similar sequence homology to the protein coding gene PTEN, functioned as a sponge for ncRNA repressors of PTEN[80][Figure 2A]. Specifically, PTENP1 binds a number of miRNAs, such as miR-21, causing disruption of cognate miR-21-PTEN base-pairing in cells[24,80,81]. When sufficient levels of PTENP1 are present, miR-21 is sequestered by the PTENP1 pseudogene which contains homologous miR-21 binding sites similar to PTEN. This results in the elevation of PTEN transcript levels, thereby promoting a tumor suppressive phenotype as PTEN inhibits the PI3K/AKT cell survival pathway[82-84]. However, one can imagine a ping-pong effect, whereby the ratio of PTENP1-PTEN abundance changes and levels of the PTEN transcript becomes more abundant. This results in miR-21 preferentially binding to PTEN causing a de-repression of the PTENP1 pseudogene. Under normal cellular conditions one can image a balanced scenario whereby miR-21 binds to either PTENP1 or PTEN in a 1:1 stoichiometric relationship. However, under certain chronic disorders, the over-abundance of any individual pseudogene can disrupt this balance, promoting inappropriate expression of transcripts that support either pro-proliferative or pro-survival signalling pathways (i.e., via the repression of PTEN transcripts)[80,85]. Therefore, further elucidating the mechanisms of ceRNA networks specific to metastasis are warranted and requires additional study.
Studies by Karreth et al.[86] have investigated these ncRNA interactions on a genome-wide level, and found that the ceRNA hypothesis can be applied to any number of ncRNAs that have the capacity for Watson-Crick base-pairing with another RNA molecule, either coding or non-coding. As an example, miRNAs can bind to and promote the decay of certain lncRNAs as these transcripts also contain a 3’UTR that in many cases harbor similar sequence motifs of the neighboring mRNA transcripts (i.e., lincRNA-p21 and CDKN1A) [Table 2]. Many of the mechanisms that facilitate miRNA-lncRNA interactions are similar to those that regulate miRNA-mRNA interactions. For instance, let-7 post-transcriptionally represses the RAS and HMGA2 oncogenes in epithelial tumors, altering the metastatic potential of these cells[87-90]. However, let-7 also binds to well established oncogenic lncRNAs such as H19 and HOTAIR[91], which promotes post-transcriptional repression of gene targets via AGO2-mediated lncRNA degradation. let-7 also reduces lncRNA levels through a separate mechanism of RNA decay by recruiting HuR binding proteins to AU-rich regions of the targeted ncRNA transcript. Other examples of lncRNAs that are regulated via the ceRNA hypothesis include interactions with lncRNA sponges. For instance, HULC and lncRNA-ATB can bind miR-372 and miR-200 respectively, but does not result in the degradation of the lncRNA[92,93]. Rather, miR-372 binding to HULC precludes miR-372 binding to bona fide mRNA targets such as LATS2[94]. This sponging phenotype of removing an inhibitor of LATS2 expression is relevant as LATS2 itself is a tumor suppressor. Therefore, HULC along with a number of other lncRNAs function as sponges or decoys that operate together to modulate the ncRNA network important for the manifestation of a particular cellular phenotype.
lncRNA-RNA associations involved in metastatic signaling cascades
| lncRNA | Interacting ncRNA | Mechanism of action | Cancer type | Phenotype | References |
|---|---|---|---|---|---|
| MALAT1 | miR-9 | miR-9 downregulation of MALAT1 | Osteosarcoma | Reduced proliferation and colony formation | [216] |
| miR-1 | competitive binding between miR-1, MALAT1, and Cdc42 | Breast cancer | Enhanced migration and invasion | [217] | |
| miR-22-3p | competitive binding between miR-22-3p, CXCR2, and MALAT1 | Sarcomas | Regulates angiogenesis | [218] | |
| HOTAIR | miR-545 | Feedback mechanism between HOTAIR, miR-545, and EGFR | Gastric cancer | Promotes EGFR-induced proliferation | [219] |
| miR-148a | HOTAIR is a miR-148a sponge regulates Snail2 | Esophageal cancer | Promotes EMT expression | [220] | |
| miR-568 | HOTAIR epigenetically represses miR-568 | Breast cancer | Promotes metastasis via enhanced angiogenesis | [221] | |
| lncRNA-ATB | miR-200 | lncRNA-ATB operates as a sponge for let-7 | Liver and gastric cancer | Regulation of ZEB and EMT | [91,115] |
| miR-372 | lncRNA-ATB competes with miR-372 and LATS2 | Liver cancer | Modulates PKA signaling and energy metabolism | [92] | |
| miR-141-3p | lncRNA-ATB competes with miR-141-3p and TGF-β | Gastric cancer | Alters cell-cycle arrest and tumor growth | [222] | |
| H19 | let-7 | H19 operates as a sponge for let-7 | Pancreatic cancer | Increases HMGA2-mediated EMT | [215] |
| miR-141 | H19 operates as a sponge for miR-141 | Gastric cancer | Induces EMT through regulation of ZEB | [223] | |
| HULC | miR-675 | H19 and miR-675 compete with Igfr and Tgfb1 binding HuR | Prostate cancer | Regulated development and angiogenesis pathways | [224,225] |
| miR-372 | lncRNA-ATB competes with miR-372 and LATS2 | Liver cancer | Modulates PKA signaling and energy metabolism | [92] | |
| miR-200 | HULC modulates Myc expression via miR-200a as sponge | CML | Inhibits tumor growth | [226,227] | |
| lincRNA-ROR | miR-205 | lincRNA-ROR sponges miR-205 | Breast cancer | Induces EMT through regulation of ZEB | [228] |
lncRNAs modulate cell signaling pathways
In bacteria and yeast systems researchers observed that lncRNAs associate with protein modules localized to the cellular membrane[95], indicating lncRNAs are not only present within the cytoplasm, but also operate within functionally discrete cytoplasmic compartments. The result of these lncRNA-chaperone protein interactions is the modulation of cell signaling networks through the activation or inhibition of a particular receptor tyrosine kinase (RTK) via the recruitment of cytoplasmic kinases or phosphatases. For instance, in eukaryotic systems, Uchl1 codes for an important enzyme specifically expressed within dopaminergic neurons, and the activity of this protein is regulated by an antisense SINEB2 element as well as an antisense transcript AS-Uchl1[96,97]. Under conditions of metabolic stress, such rapamycin treatment, AS-Uchl1 subcellular localization transitions from being primarily nuclear in abundance towards cytoplasmic enrichment, with discrete foci detectable by FISH at active polysomes due to the 5’ cap-independent translation of Uch11.
In another scenario, lnc-DC is a lncRNA expressed within dendritic cells, and is a crucial component for the activation of STAT3 signaling. This modulation of STAT3 activity occurs because lnc-DC binds to SHP1-containing protein foci, preventing SHP1-STAT3 interactions, and, in turn, allowing for phosphorylation of STAT3 at residue tyrosine-705 by a number of kinases[98]. This implies that lncRNAs function as scaffolds that recruit cytoplasmic enzymes (i.e., ubiquitinases, or kinases) essential in mediating post-translational modifications of cytoplasmic proteins. These observations also raise questions as to whether lncRNAs can recruit adaptor proteins such as GRB2 to the vicinity of the carboxy-termini of membrane-bound receptors containing SH2 domains, which are responsible for the direct modulation of RTK-mediated cell signaling cascades. These observations also support an earlier hypothesis from studies on RNA viruses, concerning RNA-lipid interactions, specifically those with charged moieties including phosphatidylcholine (PC) and phosphatidylserine (PS), are crucial in supporting life[99-102]. Further work is warranted to elucidate the complexities of these lncRNAs that operate as trans regulators of cell signaling pathways.
Role for ncRNAs in cancer metastasis
The role of ncRNAs in the progression of the metastatic cascade has gained interest over the past decade. Small nucleolar RNAs (snoRNAs) for instance regulate the presence of ribosomal RNA (rRNA) modifications important in modulating a number of cellular phenotypes. snoRNAs are essential modulators of pre-rRNA processing through formation of a 10-21 nt RNA duplex around a specific base-pair modification[103-105]. These modifications direct the snoRNA complex to the location of enzymatic cleavage of A-sites on the pre-rRNA molecule resulting in liberation from the rRNA processing complex. snoRNAs are also important in the regulation of the spliceosome complex and the splicing of introns across a number of RNA molecules, including mRNAs, lncRNAs, and rRNAs[106,107]. snoRNAs can also regulate mRNA molecules at single nt resolution, mostly via methylation of adenosines (i.e., m6A) that alter the post-transcriptional processing of those modified mRNAs, or via 2’-O-ribose methylation of the spliceosomal machinery. Finally, snoRNAs are expressed from independent transcripts indicating each snoRNA gene is potentially regulated in a spatial and temporal pattern, and implies the regulatory mechanisms controlling a process such as RNA splicing is highly cell-context specific[108]. For instance, snoRNAU50 is responsible for methylating residue C248 on the 28S rRNA subunit, and has been further implicated in supporting a tumor-suppressor phenotype in breast and prostate cancer[109]. However, SNORD26 and SNORD30 are important regulators of rRNA processing, and are expressed at higher abundance in metastatic prostate tumor samples, as compared to those having low Gleason scores, presumably to support the increased demand for protein synthesis required during tumorigenesis[17,110]. These studies support the notion that ncRNAs such as snoRNAs operate in a cell-context dependent manner, and therefore the continued investigation of specific ncRNAs responsible for modulating RNA splicing events, or the addition of RNA modifications that support a favorable cellular environment for processes such as metastasis to occur, are important. Additional ncRNAs such as miRNAs also have a well described role within the metastatic cascade yet are beyond the scope of this review. Herein, we highlight the role lncRNAs play in promoting the metastatic cascade across a variety of cancer models[45,111,112].
lncRNAs have been reported to control one of the most well described processes within the metastatic cascade, namely the loss of E-cadherin expression on epithelial cells. Loss of E-cadherin expression is crucial in ensuring proper epithelial cell-cell adhesion is maintained, as cell-cell connections are present so as to support a state of quiescence within differentiated epithelial tissue[113]. Evidence supporting this notion includes studies assessing the mutational inactivation of E-cadherin or the elucidation of mechanisms underlying the post-transcriptional repression of E-cadherin mRNA levels[114]. Taken together, observations by numerous investigators support a widely accepted hypothesis that loss of cellular polarity through the disruption of cell-to-cell or cell-to-extracellular matrix (ECM) contacts is required for the initiation of metastasis.
Specific lncRNAs crucial in controlling E-cadherin abundance include FEZF1-AS1, which is found to be dysregulated in non-small cell lung cancer (NSCLC) samples, as compared to adjacent normal tissue samples[115]. FEZF1-AS1 is also highly expressed in poorly differentiated tumor tissues as well from as those of advanced tumor stage. FEZF1-AS1 abrogates the expression of E-cadherin by directly competing for LSD1 binding, which disrupts the required association between E-cadherin and the LSD1/EZH2 complex necessary for reducing turnover of the E-cadherin molecule itself. Therefore, lncRNAs operate not only as sponges or decoys that modulate the RNA network within a cell, but also as disruptors of cytoplasmic protein complexes essential in maintaining cellular polarity.
Another example of a lncRNA that regulates E-cadherin abundance includes lncRNA-ATB, which promotes the invasion of colorectal cancer cells after TGF-β activation. This is a relevant mechanism to study, as lncRNA-ATB harbors clinicopathologic significance, and correlates with tumor stage, as well as the presence of metastatic foci within the sentinel lymph node and/or at distant organ sites. Furthermore, lncRNA-ATB associates with reduced overall- and disease-free survival within colon cancer patients[116], and is elevated in the serum of patients post-surgery, indicating lncRNAs are present in circulating biofluids and function as biomarkers for tumor progression. Overall, with the advent of genome-wide transcriptomic studies, consortiums such as ENCODE[117] and TCGA[27] have amassed a vast array of information that investigators can utilize to elucidate how a particular lncRNA can modulate a series of RNA interaction networks involved in the attenuation of metastatic phenotypes within a cell. Given this effort, there are a number of newly identified lncRNAs strongly associated with metastasis that have the potential to be clinically relevant readouts for this biological process. Here, we report on several lncRNAs that play an important role in the metastatic process[118-120].
The new linc’s on the block
Since 2012, the number of studies highlighting lncRNA involvement within the metastatic process has increased nearly 20-fold to approximately 200 manuscripts being reported in Pubmed.gov this year. Half of these papers discuss how particular lncRNAs regulate the biochemical steps crucial for the initiation and maintenance of metastatic dissemination. These new lncRNAs are intriguing entities to study, as they have putative tumorigenic activity across a number of epithelial tumors, and are expressed at levels sufficient enough for investigators to perform both gain- and loss-of-function studies, and to assess the phenotypes that result upon lncRNA dysregulation.
Elucidating the role of these lncRNAs could further illuminate our understanding of the regulatory processes involved in the initiation of cellular depolarization and motility, as well as the crucial genetic factors required for metastatic dissemination. Below, we highlight examples of a few lncRNAs that regulate important cellular activities in epithelial tumors, which could be utilized for the development of new therapeutics for patients with metastatic disease.
H19
H19 is a 2.3-kb oncofetal lncRNA gene derived from the IGF2 locus important in regulating cellular differentiation programs during development, including maternal imprinting[120]. While H19 is expressed from only one parental allele, robust levels of H19 are present during embryonic development, which is rapidly downregulated postnatally[121-125]. Improper H19 gene dosage compensation due to the lack of maternal imprinting results in embryonic lethality in mice, associates with certain clinical manifestations of those with Beckwith-Wiedemann syndrome, and correlates with an increased risk of developing Wilms tumor of the kidney[126-129]. H19 is also highly expressed in a number of tumors, and supports metastases by antagonizing ncRNAs and epigenetic regulators, such as chromatin modifiers crucial in maintaining epithelial polarity[130]. A recent study indicated that a single nucleotide polymorphism (SNP), rs2107425 located within an intron of the H19 gene, was associated with reduced metastatic free survival. This SNP does not affect the abundance of H19 in breast cancer patients, as compared to those not harboring the rs2107425 variant. Instead, this SNP alters the activity of H19 either by preventing binding to a cognate ncRNA or RBP responsible for modulating metastatic processes, or by promoting an alternative splicing event resulting in the modulation of the ceRNA network[131].
H19 also has a direct role in regulating the cellular processes of invasion and angiogenesis crucial for the progression of metastatic disease. For instance, H19 associates with the TF enhancer of zeste homolog 2 (EZH2) in turn downregulating the expression of gatekeeper genes such as E-cadherin and adenoma polyposis coli (APC)[131,132]. H19 also supports constitutive WNT signaling by inhibiting the activity the WNT-antagonist Nkd1. Given Nkd1 inhibits WNT activity, it is plausible H19 is a crucial regulator of an autoregulatory feedback loop important in preventing the stochastic expression of WNT family members. The importance of WNT signaling as regulators of metastatic progression are discussed later in this review. However, Nkd1 itself is a specific regulator of clock and is regulated in an oscillatory manner by a number of factors. This also implies H19 synergizes with WNT/NKD1 signaling to regulate the circadian rhythm pathways essential for proper vertebrate embryogenesis, but also the molecular clock genes that provide important spatial information for the inappropriate re-activation of embryonic genes that induce tumorigenic processes such as proliferation, invasion, angiogenesis, as well as EMT[133,134].
Understanding the regulation of H19 is important as certain types of cancer are dependent upon H19. In fact, BC-819 is an approach utilizing a plasmid expression system coding for diphtheria toxin under the control of an H19 regulatory sequence. Intratumoral injection of BC-819 in vivo as well as intraperitoneal (IP) injection of the compound in ovarian cancer patients is undergoing phase I/II clinical trials and shows promise at extending survival rates by reducing tumor burden[135-137]. Additional clinical trials include the ectopic expression of BC-819 via intravesical instillation in bladder cancer patients, and BC-819 vaccination in combination with gemcitabine for those with pancreatic adenocarcinoma. While both trials show promise as an effective approach to deliver lncRNAs in cancer patients[135-137], it will be interesting to determine the specificity of BC-819 in mitigating the number of metastatic foci detectable in these patients or if this therapy extends tumor latency.
SPRY4 intronic transcript 1
Recently, SPRY4 intronic transcript 1 (SPRY4-IT1) was reported as a novel lncRNA crucial in regulating the initiation and progression of EMT across a number of epithelial carcinomas[138]. SPRY4-IT1 is transcribed from an intronic region within the SPRY4 gene and is approximately 708bp in length. SPRY4-IT1 is unique in that this lncRNA contains several known hairpin structures that associate with particular RBPs[139]. Therefore, investigators have some notion regarding the mechanisms by which SPRY4-IT1 controls cellular processes important in supporting a metastatic phenotype and includes altering the expression of regulatory genes such as MCM2, XIAP, LPIN2[138]. Functional studies indicate that aberrant expression of SPRY4-IT1 also modulates the migratory and invasive capacities across a number of in vitro cancer models and does this in part by regulating DNA repair genes, such as MDM2 and CDK1.
SPRY4-IT1 also controls the process of EMT through the modulation of intermediate filament proteins, such as fibronectin and vimentin, resulting in the fine-tuning of the molecular inputs initiated by Snail and TGF-β localization and activity. Specifically, in esophageal squamous cell carcinoma (ESCC), the overexpression of SPRY4-IT1 disrupts the nuclear localization of Snail, and facilitates TGF-β-induced EMT[140-142]. Mechanistically, it is still unclear as to how SPRY4-IT1 reduces the expression of epithelial cadherins (i.e., E-cadherin), while subsequently promoting the expression of neuronal or mesenchymal cadherins (i.e., N-cadherin). SPRY4-IT1 is known to modulate gene activity via dampening global H3K27me3 distribution, which is not in of itself not biologically informative; however, SPRY4-IT1 can induce H3K27me3 methylation of the EZH2 gene promoter, and subsequently promote the repression of E-cadherin gene expression[143]. This post-transcriptional chaperone activity of chromatin modifying complexes to particular gene loci is a typical feature of lncRNAs and is crucial in dampening unwarranted transcripts during cellular development.
It would also be interesting to determine if SPRY4-IT1 interacted with a selective subset of miRNAs that supported the development of angiogenesis. For instance, miR-126 inhibits tumor growth and results in decreased micro-vessel density in cervical cancer[144,145]. SPRTY-related proteins, such as SPRED1 have been reported to bind to miR-126 and control tumor neo-angiogenesis, as well. Therefore SPRY4-IT1 may be a crucial component of the ncRNA network responsible for tumor neo-angiogenesis.
Highly upregulated in liver cancer
Highly upregulated in liver cancer (HULC) was first identified in hepatocellular cancer and is highly expressed in liver cancer, as well as in a number of carcinomas that metastasize to the liver, including colon and breast cancer[146,147]. Two recent studies suggest that HULC promotes angiogenesis as well as neo-angiogenesis, essential processes in the progression of metastasis as micro-metastatic lesions require an oxygen-rich environment to meet the demand of tumor growth within an hypoxic environment. For instance, Zhao et al.[148] found that overexpression of HULC in hepatocellular cancer cells results in an increase in the pro-angiogenic factor SPHK1. Specifically, HULC functions by sequestering miR-107, a bona fide target of which is the TF E2F1. The derepression of E2F1 results in the enhanced transcription of SPHK1, and by extension enhanced rates of vessel tube formation as well as increased tumor burden in a murine xenograft model. This HULC-specific upregulation of angiogenic processes was further tested in a chicken chorioallantoic membrane (CAM) assay, whereby condition medium from HULC overexpressing cells promoted increased growth of vessels within the chicken embryo.
Additional mechanisms by which HULC induces angiogenesis includes the sponging of miR-372 away from genes important in modulating the growth and survival of cancer cells. Additionally, HULC itself is transcriptionally upregulated by pro-metastatic growth factors, receptors, and RBPs including IGF2 mRNA-binding protein 1 (IGF2BP1), as well as members of the protein kinase A (PKA) signalling pathway[146,149]. Going forward, HULC shows promise as a therapeutic target for patients with metastatic disease, and therefore further investigation is warranted.
Estrogen receptor regulated lincRNA 01
The role for lncRNAs regulating hormone-signaling pathways related to the metastatic cascade are less well understood. In general, 17β-estradiol is known to regulate the activity and abundance of TGFβ signaling[150,151], as well as modulate the levels of E-cadherin[152]. Together, 17β-estradiol, and moreover active ERα signaling are crucial in maintaining an epithelial phenotype by suppressing the pathways associated with EMT. 17β-estradiol/ERα signaling also controls the activity of certain ncRNAs, such as the miR-200 family of miRNAs, which regulates EMT promoting TF regulators such as ZEB1 and Smad interacting protein 1 (SIP1)[31]. This indicates steroid hormone signaling pathways can modulate ncRNA networks responsible for tumorigenesis and metastatic dissemination. For instance, our group identified lncRNA estrogen receptor regulated lincRNA 01 (ERRLR01) as a prognostic biomarker in breast cancer, which is regulated by ERα activity in breast cancer tumors. Specifically, ERRLR01 is highly expressed in triple-negative breast cancers, yet not in samples derived from patients with ERα+ tumors. Follow up experiments indicated 17β-estradiol also altered the levels of ERRLR01 in ERα+ cells lines (i.e., MCF-7 and T47D)[25,153].
Given 17β-estradiol is a crucial regulator of EMT[28], we surmise ERRLR01 and other lncRNAs are crucial mediators of the metastatic cascade. Another example of a hormone-sensitive lncRNA is linc00461, which modulates the activity of CREB, a known 17β-estradiol regulated TF. Interestingly, linc00461 interacts with miR-9 as a sponge releasing miR-9 from its cognate mRNA targets, thereby altering the activity of CREB, and subsequently modulating the proliferation and migration of glioma cells[154]. linc00461 also regulates tumorigenic and metastatic phenotypes in melanoma cells[155], therefore further work elucidating the mechanism of action for linc00461 is warranted.
Colon cancer associated transcript 2
The lncRNA colon cancer associated transcript 2 (CCAT2) was first discovered via genome-wide SNP-association studies whereby investigators determined if particular genomic variants associated with cancer incidence[156]. Previously many SNPs remain understudied because they occur within the ncRNA region of the genome. With our current understanding of the ncRNA landscape, new variants are being reassessed for functional significance in cancer. SNP, rs6983267, was of particular interest as this variant maps to the 8q24 region of the genome, which correlates with higher incidences of a number of epithelial tumors, including colorectal, prostate, ovarian, and inflammatory breast cancer[157]. Subsequent studies indicate CCAT2 levels are expressed at higher frequencies in tumor samples from colorectal cancer (CRC) patients with metastatic disease[111,156,158,159]. CCAT2 is also highly expressed in small cell lung cancer (SCLC) samples and is highly correlated with poor prognosis, as well as the presence of metastasis, signifying CCAT2 is an independent prognostic biomarker and/or therapeutic target for a disease with limited therapeutic options.
The regulatory mechanisms by which CCAT2 controls the progression of metastatic events is still unclear. Gain- or loss-of-function studies demonstrate that CCAT2 can modulate the proliferation and invasion potential of cancer cells in vitro, as well as the number of micro-metastatic lesions at distant organ sites utilizing murine xenograft models. However, the only reported cellular mechanism of action by which CCAT2 alters the metastatic potential of cells involves WNT signaling. The WNT gene family are crucial regulators of metastatic progression as WNT coupling to Frizzled receptors on the cell surface allows for the release of β-catenin from the GSK-3β ubiquitination complex[160,161]. β-catenin then enters the nucleus and operates as a transcriptional co-activator along with TCF/LEF, which together promotes the transcription of genes supporting metastatic progression. Here, CCAT2 overexpressing cell lines harbor increased WNT activity, while siRNAs directed towards CCAT2 reduced both the nuclear and cytoplasmic abundance of β-catenin, which in turn resulted in reduced CCND1 and MYC protein expression[162-168]. Moreover, the phenotypes observed under CCAT2 knockdown conditions operated synergistically with small molecule WNT inhibitor, FH-535. These studies indicate CCAT2 imparts a specific regulatory function through the augmentation of the WNT signaling pathway, and as such, contributes to a pro-metastatic phenotype.
Myocardial infarction associated transcript
The lncRNA myocardial infarction associated transcript (MIAT) has been linked to several chronic disorders including myocardial infarction[169,170], paranoid schizophrenia[171], and neuroendocrine-derived prostate cancers[110]. MIAT interacts with a number of epigenetic modifiers in neuronal crest cells including PRC1/2 and ZEB1 that when altered results in the modified migratory capacities of these cell lines[172]. MIAT is also disrupted in a number of cancer cell lines, and is expressed at significantly lower levels in grade I-II breast tumors, as compared to those considered high grade III-IV tumors[110]. MIAT also modulates the invasive capacities of breast cancer cells by mediating the ncRNA interaction networks between ZEB1 and certain miRNAs, such as miR-150 and miR-29[173]. For instance, knockdown of MIAT promotes the expression of miR-150, yet also results in the reduction of miR-29 levels. This results in breast tumor cell lines transitioning from a pro-proliferative state towards a more quiescent yet migratory phenotype[72,174,175]. There are two mechanisms by which MIAT could alter these miRNA interaction networks. The first is that MIAT functions as a ceRNA and operates as a sponge for miR-150, reducing the bioavailability of miR-150 to interact with cognate mRNA transcripts such as ZEB1. The second is that MIAT operates as a chaperone or scaffold that recruits co-repressor complexes to the promoter of the MIR150 host gene, subsequently reducing the transcriptional output of miR-150. While both scenarios may not be mutually exclusive from one another, these regulatory interactions reinforce the notion that EMT is a highly controlled cellular program responsible for modulating the expression of E-cadherin, which is initiated by several key transcriptional repressors including ZEB1[176,177]. Interestingly, the resulting consequence of the MIAT-miR-150 interaction is the increased expression of ZEB1 and, in turn, a pro-metastatic phenotype, through the transcriptional upregulation of the MIR29 host gene. miR-29 promotes the invasive capabilities of cells by diverting the energy demand required for cellular proliferation and redirecting those energies towards signaling pathways that encourage motility and invasion[178,179]. This occurs in part by modulating the expression of cell-cycle checkpoint genes such as CDKN2A. Therefore, MIAT represents a characteristic example of how lncRNAs can modulate the activity of RNA-RNA interactions by controlling the bioavailability of other ncRNAs, in turn, reprograming cellular signaling cascades to support a pro-metastatic phenotype.
BMP/OP-responsive gene
Recently, the lncRNA BMP/OP-responsive gene (BORG) was identified to play a vital role in augmenting proliferation and survival cues within breast cancer cells[71,180]. Specifically, BORG interacts with the TRIM28 TF, which modulates the transcriptional co-repression of Cdkn1a and Gadd45a[181,182]. The presence of this BORG-TRIM28 binding complex is also linked with shorter tumor latency within breast cancer patients and correlates with a faster outgrowth of cancer cells in 3D culture systems. TRIM28 can also function as a transcriptional activator or repressor depending upon the chromatin architecture or extent of heterochromatization within the nucleus. BORG localizes predominantly to the nucleus and has a unique function as it reinforces the repressive actions associated with TRIM28. As an example, repression of BORG in metastatic D2.A1 breast cancer cells prevents migratory outgrowths within 3D matrigel culture systems, as well as the abundance of micro-metastatic colonies in lung tissue utilizing an invasive breast cancer transplant model[180]. Furthermore, in aggressive metastatic breast cancer the expression of BORG is higher as compared to samples derived from non-malignant mammary tissues. Therefore, BORG clearly modulates the invasive capacities of breast cancer cells.
While RNA immunoprecipitation (RIP) experiments indicate that TRIM28 in fact requires BORG for binding to specific gene promoter regions, such as those neighboring Cdkn1a and Gadd45a[180], it is still unclear as to how BORG modulates TRIM28 binding to specific chromatin regions that in turn modulates a metastatic phenotype. One assumption is that specific sequence regions of BORG, outside the identified TRIM28 binding sequence, interacts with TRIM28 or additional TRIM domain-containing TFs through stacking interactions. These lncRNA structure-based interactions, mediate recruitment of the TRIM28 transcriptional protein complex to the proximal promoters of genes such as Cdkn1a and Gadd45a. BORG also confers a unique transcriptional signature that is enriched for KRAS signaling, as compared to non-metastatic D2.OR breast cancer cells. Further studies elucidating the role of BORG in human breast cancer cells, as well as the regulatory role within the metastatic process, are warranted.
Prostate cancer associated transcript 1
Through genome wide RNA sequencing experiments, Prensner et al.[183] identified prostate cancer associated transcript 1 (PCAT1) as a lncRNA highly upregulated in metastatic prostate cancer samples, as well as those considered high grade (i.e., stage II-IV). Upon knockdown of PCAT1 in prostate cancer cell lines, Prensner et al.[183] identified 370 genes expressed differentially, many of which were associated with cell-cycle progression and mitosis, as well as cytoskeleton and microtubule regulation. Knockdown studies indicated that loss of PCAT1 resulted in an approximate 25% reduction in cellular proliferation, though the mechanism by which PCAT1 promotes an invasive phenotype is still unclear. One possibility may be due to the involvement of PCAT1 in the homologous recombination (HR) repair pathway. One can surmise, for instance, that as epithelial progenitor cells proliferate, the acquisition of successive mutations within the genome across daughter generations provides an opportunity for such cells to undergo a process such as EMT[12]. Interestingly, PCAT1 is inversely correlated with BRCA2 expression in LNCaP cells, while the knockdown of PCAT1 resulted in the upregulation of BRCA2, a crucial component of the DNA repair pathway[184,185]. Moreover, PCAT1 overexpression alters the formation of RAD51 and ɣ-H2aX foci after radiation-induced DNA damage, while naturally occurring polymorphisms within the genome, such as rs7463708, can promote an enhanced proliferative and migratory state within prostate cancer cell lines[186]. Therefore, it is entirely plausible that the reduction in chromosome stability via enhanced PCAT1 activity supports not only a pro-tumorigenic state, but a pro-metastatic phenotype as well. Separating these two distinct yet equally important mechanisms will be crucial in developing novel therapeutics to treat those with advanced prostate cancer.
Conclusion
Overall, lncRNAs play a multifaceted role in controlling the ncRNA network, which is vitally important throughout embryogenesis and vertebrate development. Here we discussed the ways in which lncRNAs can function as metastatic regulators, primarily by controlling epigenetic mechanisms, such as the recruitment of co-repressor complexes including PRC1/2, as well as co-transcriptional complexes such as CREB/REST to specific chromatin regions. Therefore, lncRNAs represent a unique class of ncRNA that operate as scaffolds to bring specific chromosomal foci into proximity with epigenetic regulators and chromatin modifiers. lncRNAs also control the appropriate expression of the DNA methylation machinery such as DNMT1, and function as competitive binding partners for other ncRNAs with complementary sequences. As such, lncRNAs serve as potent disruptors of conserved RNA-RNA regulatory networks.
Interestingly, lncRNA sequences are not highly conserved across species, however lncRNAs harbor a conserved positional synteny that is linked with the regulatory function of that specific lncRNA. This presents a unique challenge for the lncRNA field in that determining the importance of a lncRNA molecule found to be differently expressed under certain experimental conditions cannot be further studied by assessing the conservation of the sequence. Investigators will require a more nuanced approach in studying the landscape of the surrounding genomic architecture, the proximity of certain DNA response elements, and if specific protein coding genes flank the lncRNA, while also keeping in mind the state of the surrounding chromatin architecture and determining if the DNA region is highly hetero- or eu-chromatinized [Figure 2C].
This also means that the function of a particular lncRNA using a mouse model of metastasis for instance, does not always imply that the mechanism of action of those lncRNAs function similarly in human cells. For instance, the use of genetically engineered mouse models (GEMMs) that have a propensity to develop metastasis may provide some useful information yet may not provide a complete picture regarding the mechanisms by which a specific lncRNA promotes metastasis in human systems. Therefore, additional technologies will be required to assess the functionality of metastatic-specific lncRNAs. For instance, the use of humanized mouse models, 3D culture systems, and use of conditionally reprogrammed cells from human tissue, all will aid investigators in determining the bona fide relevance of functionally conserved lncRNAs.
In many cases lncRNAs are expressed at lower abundance than cytoplasmic mRNA, thereby making it difficult to assess whether lncRNAs are functionally relevant, or present as a result of leaky transcriptional activity. As an example, lncRNAs can regulate the processing of nascent transcripts generated from RNA polII-based transcription. These lncRNAs may only number a few copies in the cell at any given time yet can bind in a 1:1 stoichiometric relationship with the nascent mRNA altering the stability of the newly synthesized RNA molecule. Technologies such as global run on sequencing[187], which is a sensitive and high throughput type of nuclear-run on assay, have been developed to specifically determine the relevant abundance of a particular lncRNA binding these nascent transcripts. Additionally, techniques such as high-throughput sequencing of RNAs isolated by crosslinking immunoprecipitation (CLIP)[188] and cross-linking, ligation and sequencing of hybrids (CLASH)[189] have been utilized to gauge the abundance, enrichment, and/or composition of composition of ncRNAs within particular RNA-RBP cytoplasmic complexes. As sequencing technology develops and the cost to perform these analyses decrease, the utilization of these biochemical approaches coupled with these high-throughput sequencing methods will pave the way for new discoveries regarding lncRNA function.
Despite these challenges, it is clear that lncRNAs play a crucial role in driving a metastatic phenotype, and in particular regulate the initiating steps of metastasis such as EMT. Given EMT is the process of cell fate switching, or reactivation of embryogenic programs that convert epithelia cells to those harboring a mesenchymal phenotype, the continued approach of utilizing reductionist-based investigations within well-defined model systems will help in elucidating the mechanisms by which individual lncRNAs regulate the underlying biology of metastasis. Given the advances in sequencing technology as well as a renewed scientific interest in lncRNA biology, the number of publication discussing the role of lncRNAs in metastasis will most likely double in the next year. The continued demand for reliable biomarkers of metastasis will also fuel research towards the development of prognostic and predictive indicators for patients with high grade tumors harboring metastatic dissemination. In conclusion, the lncRNA field is certainly in its infancy, yet is considered to be the wild-west of the post-genomic era and has the potential to unlock the key to some of the most prevalent challenges associated with treating patients with metastatic disease.
Declarations
AcknowledgmentsWe thank Kimberly N. Kelly at BioCT Innovation Commons, as well as Drs. Garret Barr and Ann Yezerski Gilmor at King’s College for providing the resources and office space required for the writing of this manuscript.
Authors’ contributionsConception and design: Adams BD, Reed IG, Alexander A, Willetts L, Habibian M
Financial support: Adams BD
Literature search, manuscript preparation and writing: all authors
Manuscript editing: Adams BD, Reed IG, Willetts L
Manuscript review: all authors
Financial support and sponsorshipThis work was supported by The Brain Institute of America Foundation.
Conflicts of interestBrian D. Adams is President/CEO of The Brain Institute of America and holds patent interests with AUM LifeTech. Authors have no other conflicts of interest to disclose. The authors declare that the research was conducted in the absence of any commercial or financial activities.
Patient consentNot applicable.
Ethics approvalNot applicable.
Copyright© The Author(s) 2018.
REFERENCES
1. DeSantis C, Siegel R, Bandi P, Jemal A. Breast cancer statistics, 2011. CA Cancer J Clin 2011;61:409-18.
3. Li J, Meng H, Bai Y, Wang K. Regulation of lncRNA and its role in cancer metastasis. Oncol Res 2016;23:205-17.
5. Chiang SP, Cabrera RM, Segall JE. Tumor cell intravasation. Am J Physiol Cell Physiol 2016;311:C1-14.
6. Simpson CD, Anyiwe K, Schimmer AD. Anoikis resistance and tumor metastasis. Cancer Lett 2008;272:177-85.
7. Irmisch A, Huelsken J. Metastasis: new insights into organ-specific extravasation and metastatic niches. Exp Cell Res 2013;319:1604-10.
8. Ladetto M, Brüggemann M, Monitillo L, Ferrero S, Pepin F, Drandi D, Barbero D, Palumbo A, Passera R, Boccadoro M, Ritgen M, Gökbuget N, Zheng J, Carlton V, Trautmann H, Faham M, Pott C. Next-generation sequencing and real-time quantitative PCR for minimal residual disease detection in B-cell disorders. Leukemia 2014;28:1299-307.
10. Tsuji T, Ibaragi S, Shima K, Hu MG, Katsurano M, Sasaki A, Hu GF. Epithelial-mesenchymal transition induced by growth suppressor p12CDK2-AP1 promotes tumor cell local invasion but suppresses distant colony growth. Cancer Res 2008;68:10377-86.
11. Klymkowsky MW, Savagner P. Epithelial-mesenchymal transition: a cancer researcher's conceptual friend and foe. Am J Pathol 2009;174:1588-93.
12. Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer 2009;9:265-73.
13. Barrallo-Gimeno A, Nieto MA. The Snail genes as inducers of cell movement and survival: implications in development and cancer. Development 2005;132:3151-61.
14. Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest 2009;119:1420-8.
15. Burger GA, Danen EH, Beltman JB. Deciphering epithelial-mesenchymal transition regulatory networks in cancer through computational approaches. Front Oncol 2017;7:162.
16. Spano D, Heck C, De Antonellis P, Christofori G, Zollo M. Molecular networks that regulate cancer metastasis. Semin Cancer Biol 2012;22:234-49.
17. Crea F, Clermont PL, Parolia A, Wang Y, Helgason CD. The non-coding transcriptome as a dynamic regulator of cancer metastasis. Cancer Metastasis Rev 2014;33:1-16.
18. Adams BD, Parsons C, Walker L, Zhang WC, Slack FJ. Targeting noncoding RNAs in disease. J Clin Invest 2017;127:761-71.
19. Abdul-Rahman U, Győrffy B, Adams BD. linc00673 (ERRLR01) is a prognostic indicator of overall survival in breast cancer. Transcription 2018;9:17-29.
20. Adams BD, Slack FJ. MicroRNA Signatures as Biomarkers in Cancer. In: eLS. Chichester: John Wiley & Sons Ltd; 2015.
21. Adams BD, Furneaux H, White BA. The micro-ribonucleic acid (miRNA) miR-206 targets the human estrogen receptor-alpha (ERalpha) and represses ERalpha messenger RNA and protein expression in breast cancer cell lines. Mol Endocrinol 2007;21:1132-47.
22. Metheetrairut C, Adams BD, Nallur S, Weidhaas JB, Slack FJ. cel-mir-237 and its homologue, hsa-miR-125b, modulate the cellular response to ionizing radiation. Oncogene 2017;36:512-24.
23. Lang MF, Yang S, Zhao C, Sun G, Murai K, Wu X, Wang J, Gao H, Brown CE, Liu X, Zhou J, Peng L, Rossi JJ, Shi Y. Genome-wide profiling identified a set of miRNAs that are differentially expressed in glioblastoma stem cells and normal neural stem cells. PLoS One 2012;7:e36248.
24. Conti A, Aguennouz M, La Torre D, Tomasello C, Cardali S, Angileri FF, Maio F, Cama A, Germanò A, Vita G, Tomasello F. miR-21 and 221 upregulation and miR-181b downregulation in human grade II-IV astrocytic tumors. J Neurooncol 2009;93:325-32.
27. Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumors. Nature 2012;490:61-70.
28. Guttilla IK, Adams BD, White BA. ERα, microRNAs, and the epithelial–mesenchymal transition in breast cancer. Trends Endocrinol Metab 2012;23:73-82.
29. Adams B, Guttilla IK, White B. Involvement of microRNAs in breast cancer. Semin Reprod Med 2008;26:522-36.
30. Kondaveeti Y, Guttilla Reed IK, White BA. Epithelial-mesenchymal transition induces similar metabolic alterations in two independent breast cancer cell lines. Cancer Lett 2015;364:44-58.
31. Peter ME. Let-7 and miR-200 microRNAs: guardians against pluripotency and cancer progression. Cell Cycle 2009;8:843-52.
32. Singh T, Adams BD. The regulatory role of miRNAs on VDR in breast cancer. Transcription 2017;8:232-41.
34. Adams BD, Wali VB, Cheng CJ, Inukai S, Booth CJ, Agarwal S, Rimm DL, Győrffy B, Santarpia L, Pusztai L, Saltzman WM, Slack FJ. MiR-34a silences c-SRC to attenuate tumor growth in triple-negative breast cancer. Cancer Res 2016;76:927-39.
35. Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer 2006;6:259-69.
36. Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 2009;19:92-105.
38. Fan M, Krutilina R, Sun J, Sethuraman A, Yang CH, Wu ZH, Yue J, Pfeffer LM. Comprehensive analysis of microRNA (miRNA) targets in breast cancer cells. J Biol Chem 2013;288:27480-93.
39. Ma L, Reinhardt F, Pan E, Soutschek J, Bhat B, Marcusson EG, Teruya-Feldstein J, Bell GW, Weinberg RA. Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nat Biotechnol 2010;28:341-7.
41. Chen Y, Min L, Zhang X, Hu S, Wang B, Liu W, Wang R, Gu X, Shen W, Lv H, Zou J, Chen Y, Xu X, Chen L. Decreased miRNA-148a is associated with lymph node metastasis and poor clinical outcomes and functions as a suppressor of tumor metastasis in non-small cell lung cancer. Oncol Rep 2013;30:1832-40.
42. Jiang Z, Yin J, Fu W, Mo Y, Pan Y, Dai L, Huang H, Li S, Zhao J. miRNA 17 family regulates cisplatin-resistant and metastasis by targeting TGFbetaR2 in NSCLC. PLoS One 2014;9:e94639.
43. Zhang J, Ma L. MicroRNA control of epithelial-mesenchymal transition and metastasis. Cancer Metastasis Rev 2012;31:653-62.
47. Morris KV, Santoso S, Turner AM, Pastori C, Hawkins PG. Bidirectional transcription directs both transcriptional gene activation and suppression in human cells. PLoS Genet 2008;4:e1000258.
48. Yap KL, Li S, Mu-oz-Cabello AM, Raguz S, Zeng L, Mujtaba S, Gil J, Walsh MJ, Zhou MM. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell 2010;38:662-74.
49. Johnsson P, Ackley A, Vidarsdottir L, Lui WO, Corcoran M, Grandér D, Morris KV. A pseudogene long-noncoding-RNA network regulates PTEN transcription and translation in human cells. Nat Struct Mol Biol 2013;20:440-6.
50. Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M, Davis CA, Shiekhattar R, Gingeras TR, Hubbard TJ, Notredame C, Harrow J, Guigó R. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 2012;22:1775-89.
52. Ferrè F, Colantoni A, Helmer-Citterich M. Revealing protein-lncRNA interaction. Brief Bioinform 2016;17:106-16.
54. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013;495:333-8.
55. Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A 2006;103:2257-61.
56. Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993;75:843-54.
57. Kiss T. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J 2001;20:3617-22.
58. Kiss T. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell 2002;109:145-8.
59. López-Maury L, Marguerat S, Bähler J. Tuning gene expression to changing environments: from rapid responses to evolutionary adaptation. Nat Rev Genet 2008;9:583-93.
60. Bishop NA, Guarente L. Genetic links between diet and lifespan: shared mechanisms from yeast to humans. Nat Rev Genet 2007;8:835-44.
61. Luo S, Lu JY, Liu L, Yin Y, Chen C, Han X, Wu B, Xu R, Liu W, Yan P, Shao W, Lu Z, Li H, Na J, Tang F, Wang J, Zhang YE, Shen X. Divergent lncRNAs regulate gene expression and lineage differentiation in pluripotent cells. Cell Stem Cell 2016;18:637-52.
62. Kaneko S, Bonasio R, Salda-a-Meyer R, Yoshida T, Son J, Nishino K, Umezawa A, Reinberg D. Interactions between JARID2 and noncoding RNAs regulate PRC2 recruitment to chromatin. Mol Cell 2014;53:290-300.
63. Ghosal S, Das S, Chakrabarti J. Long noncoding RNAs: new players in the molecular mechanism for maintenance and differentiation of pluripotent stem cells. Stem Cells Dev 2013;22:2240-53.
64. Kurokawa R. Promoter-associated long noncoding RNAs repress transcription through a RNA binding protein TLS. Adv Exp Med Biol 2011;722:196-208.
65. Korzus E, Rosenfeld MG, Mayford M. CBP histone acetyltransferase activity is a critical component of memory consolidation. Neuron 2004;42:961-72.
66. Long Y, Wang X, Youmans DT, Cech TR. How do lncRNAs regulate transcription? Sci Adv 2017;3:eaao2110.
67. Ounzain S, Pedrazzini T. The promise of enhancer-associated long noncoding RNAs in cardiac regeneration. Trends Cardiovasc Med 2015;25:592-602.
68. Klattenhoff CA, Scheuermann JC, Surface LE, Surface LE, Bradley RK, Fields PA, Steinhauser ML, Ding H, Butty VL, Torrey L, Haas S, Abo R, Tabebordbar M, Lee RT, Burge CB, Boyer LA. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell 2013;152:570-83.
70. Werner A, Swan D. What are natural antisense transcripts good for? Biochem Soc Trans 2010;38:1144-9.
71. Flynn RA, Chang HY. Active chromatin and noncoding RNAs: an intimate relationship. Curr Opin Genet Dev 2012;22:172-8.
72. Uchida T, Rossignol F, Matthay MA, Mounier R, Couette S, Clottes E, Clerici C. Prolonged hypoxia differentially regulates hypoxia-inducible factor (HIF)-1alpha and HIF-2alpha expression in lung epithelial cells: implication of natural antisense HIF-1alpha. J Biol Chem 2004;279:14871-8.
73. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 2011;146:353-8.
76. Di Ruscio A, Ebralidze AK, Benoukraf T, Amabile G, Goff LA, Terragni J, Figueroa ME, De Figueiredo Pontes LL, Alberich-Jorda M, Zhang P, Wu M, D'Alò F, Melnick A, Leone G, Ebralidze KK, Pradhan S, Rinn JL, Tenen DG. DNMT1-interacting RNAs block gene-specific DNA methylation. Nature 2013;503:371-6.
77. McHugh CA, Chen CK, Chow A, Surka CF, Tran C, McDonel P, Pandya-Jones A, Blanco M, Burghard C, Moradian A, Sweredoski MJ, Shishkin AA, Su J, Lander ES, Hess S, Plath K, Guttman M. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 2015;521:232-6.
78. Diaz-Perez SV, Ferguson DO, Wang C, Csankovszki G, Wang C, Tsai SC, Dutta D, Perez V, Kim S, Eller CD, Salstrom J, Ouyang Y, Teitell MA, Kaltenboeck B, Chess A, Huang S, Marahrens Y. A deletion at the mouse Xist gene exposes trans-effects that alter the heterochromatin of the inactive X chromosome and the replication time and DNA stability of both X chromosomes. Genetics 2006;174:1115-33.
80. Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 2010;465:1033-8.
81. Bar N, Dikstein R. miR-22 forms a regulatory loop in PTEN/AKT pathway and modulates signaling kinetics. PLoS One 2010;5:e10859.
82. Wheler JJ, Atkins JT, Janku F, Moulder SL, Yelensky R, Stephens PJ, Kurzrock R. Multiple gene aberrations and breast cancer: lessons from super-responders. BMC Cancer 2015;15:442.
85. Poliseno L, Haimovic A, Christos PJ, Vega Y Saenz de Miera EC, Shapiro R, Pavlick A, Berman RS, Darvishian F, Osman I. Deletion of PTENP1 pseudogene in human melanoma. J Invest Dermatol 2011;131:2497-500.
86. Karreth FA, Tay Y, Perna D, Ala U, Tan SM, Rust AG, DeNicola G, Webster KA, Weiss D, Perez-Mancera PA, Krauthammer M, Halaban R, Provero P, Adams DJ, Tuveson DA, Pandolfi PP. In vivo identification of tumor- suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell 2011;147:382-95.
87. Yong SL, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev 2007;21:1025-30.
88. Watanabe S, Ueda Y, Akaboshi S, Hino Y, Sekita Y, Nakao M. HMGA2 maintains oncogenic RAS-induced epithelial-mesenchymal transition in human pancreatic cancer cells. Am J Pathol 2009;174:854-68.
89. Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell 2005;120:635-47.
90. Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science 2007;315:1576-9.
91. Yoon JH, Abdelmohsen K, Gorospe M. Functional interactions among microRNAs and long noncoding RNAs. Semin Cell Dev Biol 2014;34:9-14.
92. Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF, Liu F, Pan W, Wang TT, Zhou CC, Wang SB, Wang YZ, Yang Y, Yang N, Zhou WP, Yang GS, Sun SH. A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell 2014;25:666-81.
93. Wang J, Liu X, Wu H, Ni P, Gu Z, Qiao Y, Chen N, Sun F, Fan Q. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res 2010;38:5366-83.
94. Voorhoeve PM, le Sage C, Schrier M, Gillis AJ, Stoop H, Nagel R, Liu YP, van Duijse J, Drost J, Griekspoor A, Zlotorynski E, Yabuta N, De Vita G, Nojima H, Looijenga LH, Agami R. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Adv Exp Med Biol 2007;604:17-46.
95. Ala U, Karreth FA, Bosia C, Pagnani A, Taulli R, Léopold V, Tay Y, Provero P, Zecchina R, Pandolfi PP. Integrated transcriptional and competitive endogenous RNA networks are cross-regulated in permissive molecular environments. Proc Natl Acad Sci U S A 2013;110:7154-9.
96. Liu Y, Lansbury P. The UCH-L1 gene encodes two opposing enzymatic activities that affect a-synuclein degradation and Parkinson's disease susceptibility. Cell 2002;111:209-18.
97. Carrieri C, Cimatti L, Biagioli M, Beugnet A, Zucchelli S, Fedele S, Pesce E, Ferrer I, Collavin L, Santoro C, Forrest AR, Carninci P, Biffo S, Stupka E, Gustincich S. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature 2012;491:454-7.
98. Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S, Jiang Z, Xu J, Liu Q, Cao X. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science 2014;344:310-3.
99. Xu K, Nagy PD. RNA virus replication depends on enrichment of phosphatidylethanolamine at replication sites in subcellular membranes. Proc Natl Acad Sci U S A 2015;112:E1782-91.
100. Lee WM, Ahlquist P. Membrane synthesis, specific lipid requirements, and localized lipid composition changes associated with a positive-strand RNA virus RNA replication protein. J Virol 2003;77:12819-28.
101. Michanek A, Kristen N, Höök F, Nylander T, Sparr E. RNA and DNA interactions with zwitterionic and charged lipid membranes - a DSC and QCM-D study. Biochim Biophys Acta 2010;1798:829-38.
102. Chukkapalli V, Inlora J, Todd GC, Ono A. Evidence in support of RNA-mediated inhibition of phosphatidylserine-dependent HIV-1 Gag membrane binding in cells. J Virol 2013;87:7155-9.
103. Mishra RK, Eliceiri GL. Three small nucleolar RNAs that are involved in ribosomal RNA precursor processing. Proc Natl Acad Sci U S A 1997;94:4972-7.
104. Bratkovič T, Rogelj B. The many faces of small nucleolar RNAs. Biochim Biophys Acta 2014;1839:438-43.
106. Shah BN, Liu X, Correll CC. Imp3 unfolds stem structures in pre-rRNA and U3 snoRNA to form a duplex essential for small subunit processing. RNA 2013;19:1372-83.
107. Michot B, Joseph N, Mazan S, Bachellerie JP. Evolutionarily conserved structural features in the ITS2 of mammalian pre-rRNAs and potential interactions with the snoRNA U8 detected by comparative analysis of new mouse sequences. Nucleic Acids Res 1999;27:2271-82.
108. Pacilli A, Ceccarelli C, Treré D, Montanaro L. SnoRNA U50 levels are regulated by cell proliferation and rRNA transcription. Int J Mol Sci 2013;14:14923-35.
109. Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B, Antipin Y, Mitsiades N, Landers T, Dolgalev I, Major JE, Wilson M, Socci ND, Lash AE, Heguy A, Eastham JA, Scher HI, Reuter VE, Scardino PT, Sander C, Sawyers CL, Gerald WL. Integrative genomic profiling of human prostate cancer. Cancer Cell 2010;18:11-22.
110. Crea F, Venalainen E, Ci X, Cheng H, Pikor L, Parolia A, Xue H, Nur Saidy NR, Lin D, Lam W, Collins C, Wang Y. The role of epigenetics and long noncoding RNA MIAT in neuroendocrine prostate cancer. Epigenomics 2016;8:721-31.
111. Jing X, Liang H, Cui X, Han C, Hao C, Huo K. Long noncoding RNA CCAT2 can predict metastasis and a poor prognosis: a meta-analysis. Clin Chim Acta 2017;468:159-65.
112. Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, Xue C, Marinov GK, Khatun J, Williams BA, Zaleski C, Rozowsky J, Röder M, Kokocinski F, Abdelhamid RF, Alioto T, Antoshechkin I, Baer MT, Bar NS, Batut P, Bell K, Bell I, Chakrabortty S, Chen X, Chrast J, Curado J, Derrien T, Drenkow J, Dumais E, Dumais J, Duttagupta R, Falconnet E, Fastuca M, Fejes-Toth K, Ferreira P, Foissac S, Fullwood MJ, Gao H, Gonzalez D, Gordon A, Gunawardena H, Howald C, Jha S, Johnson R, Kapranov P, King B, Kingswood C, Luo OJ, Park E, Persaud K, Preall JB, Ribeca P, Risk B, Robyr D, Sammeth M, Schaffer L, See LH, Shahab A, Skancke J, Suzuki AM, Takahashi H, Tilgner H, Trout D, Walters N, Wang H, Wrobel J, Yu Y, Ruan X, Hayashizaki Y, Harrow J, Gerstein M, Hubbard T, Reymond A, Antonarakis SE, Hannon G, Giddings MC, Ruan Y, Wold B, Carninci P, Guigó R, Gingeras TR. Landscape of transcription in human cells. Nature 2012;489:101-8.
113. Park SH, Cheung LW, Wong AS, Leung PC. Estrogen regulates Snail and Slug in the down-regulation of E-cadherin and induces metastatic potential of ovarian cancer cells through estrogen receptor alpha. Mol Endocrinol 2008;22:2085-98.
114. He R, Zhang FH, Shen N. LncRNA FEZF1-AS1 enhances epithelial-mesenchymal transition (EMT) through suppressing E-cadherin and regulating WNT pathway in non-small cell lung cancer (NSCLC). Biomed Pharmacother 2017;95:331-8.
115. Yue B, Qiu S, Zhao S, Liu C, Zhang D, Yu F, Peng Z, Yan D. LncRNA-ATB mediated E-cadherin repression promotes the progression of colon cancer and predicts poor prognosis. J Gastroenterol Hepatol 2016;31:595-603.
116. ENCODE Project Consortium. The ENCODE (ENCyclopedia of DNA Elements) Project. Science 2004;306:636-40.
119. Doolittle WF. Is junk DNA bunk? A critique of ENCODE. Proc Natl Acad Sci U S A 2013;110:5294-300.
120. Thorvaldsen JL, Duran KL, Bartolomei MS. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev 1998;12:3693-702.
121. Collette J, Le Bourhis X, Adriaenssens E. Regulation of human breast cancer by the long non-coding RNA H19. Int J Mol Sci 2017;18:E2319.
122. Pachnis V, Brannan CI, Tilghman SM. The structure and expression of a novel gene activated in early mouse embryogenesis. EMBO J 1988;7:673-81.
123. Poirier F, Chan CT, Timmons PM, Robertson EJ, Evans MJ, Rigby PW. The murine H19 gene is activated during embryonic stem cell differentiation in vitro and at the time of implantation in the developing embryo. Development 1991;113:1105-14.
124. Rachmilewitz J, Goshen R, Ariel I, Schneider T, de Groot N, Hochberg A. Parental imprinting of the human H19 gene. FEBS Lett 1992;309:25-8.
125. Bartolomei MS, Webber AL, Brunkow ME, Tilghman SM. Epigenetic mechanisms underlying the imprinting of the mouse H19 gene. Genes Dev 1993;7:1663-73.
126. Sparago A, Cerrato F, Vernucci M, Ferrero GB, Silengo MC, Riccio A. Microdeletions in the human H19 DMR result in loss of IGF2 imprinting and Beckwith-Wiedemann syndrome. Nat Genet 2004;36:958-60.
127. DeBaun MR, Tucker MA. Risk of cancer during the first four years of life in children from The Beckwith-Wiedemann Syndrome Registry. J Pediatr 1998;132:398-400.
128. Brunkow ME, Tilghman SM. Ectopic expression of the H19 gene in mice causes prenatal lethality. Genes Dev 1991;5:1092-101.
129. Heery R, Finn S, Cuffe S, Gray SG. Long non-coding RNAs: key regulators of epithelial-mesenchymal transition, tumour drug resistance and cancer stem cells. Cancers (Basel) 2017;9:E38.
130. Riaz M, Berns EMJJ, Sieuwerts AM, Ruigrok-Ritstier K, de Weerd V, Groenewoud A, Uitterlinden AG, Look MP, Klijn JG, Sleijfer S, Foekens JA, Martens JW. Correlation of breast cancer susceptibility loci with patient characteristics, metastasis-free survival, and mRNA expression of the nearest genes. Breast Cancer Res Treat 2012;133:843-51.
131. Luo M, Li Z, Wang W, Zeng Y, Liu Z, Qiu J. Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. Cancer Lett 2013;333:213-21.
132. Berx G, Van Roy F. The E-cadherin/catenin complex: an important gatekeeper in breast cancer tumorigenesis and malignant progression. Breast Cancer Res 2001;3:289-93.
133. Ishikawa A, Kitajima S, Takahashi Y, Kokubo H, Kanno J, Inoue T, Saga Y. Mouse Nkd1, a Wnt antagonist, exhibits oscillatory gene expression in the PSM under the control of Notch signaling. Mech Dev 2004;121:1443-53.
134. Katoh M, Katoh M. WNT signaling pathway and stem cell signaling network. Clin Cancer Res 2007;13:4042-5.
135. Smaldone MC, Davies BJ. BC-819, a plasmid comprising the H19 gene regulatory sequences and diphtheria toxin A, for the potential targeted therapy of cancers. Curr Opin Mol Ther 2010;12:607-16.
136. Scaiewicz V, Sorin V, Fellig Y, Birman T, Mizrahi A, Galula J, Abu-Lail R, Shneider T, Ohana P, Buscail L, Hochberg A, Czerniak A. Use of H19 gene regulatory sequences in DNA-based therapy for pancreatic cancer. J Oncol 2010;2010:178174.
137. Amit D, Hochberg A. Development of targeted therapy for a broad spectrum of cancers (pancreatic cancer, ovarian cancer, glioblastoma and HCC) mediated by a double promoter plasmid expressing diphtheria toxin under the control of H19 and IGF2-P4 regulatory sequences. Int J Clin Exp Med 2012;5:296-305.
138. Li J, Chen Y, Chen Z, He A, Xie H, Zhang Q, Cai Z, Liu Y, Huang W. SPRY4-IT1: a novel oncogenic long non-coding RNA in human cancers. Tumor Biol 2017;39:1010428317711406.
139. Khaitan D, Dinger ME, Mazar J, Crawford J, Smith MA, Mattick JS, Perera RJ. The melanoma-upregulated long noncoding RNA SPRY4-IT1 modulates apoptosis and invasion. Cancer Res 2011;71:3852-62.
140. Xie M, Nie FQ, Sun M, Xia R, Liu YW, Zhou P, De W, Liu XH. Decreased long noncoding RNA SPRY4-IT1 contributing to gastric cancer cell metastasis partly via affecting epithelial-mesenchymal transition. J Transl Med 2015;13:250.
141. Cui F, Wu D, He X, Wang W, Xi J, Wang M. Long noncoding RNA SPRY4-IT1 promotes esophageal squamous cell carcinoma cell proliferation, invasion, and epithelial-mesenchymal transition. Tumour Biol 2016;37:10871-6.
142. Zhang CY, Li RK, Qi Y, Li XN, Yang Y, Liu DL, Zhao J, Zhu DY, Wu K, Zhou XD, Zhao S. Upregulation of long noncoding RNA SPRY4-IT1 promotes metastasis of esophageal squamous cell carcinoma via induction of epithelial-mesenchymal transition. Cell Biol Toxicol 2016;32:391-401.
143. Zhou M, Zhang XY, Yu X. Overexpression of the long non-coding RNA SPRY4-IT1 promotes tumor cell proliferation and invasion by activating EZH2 in hepatocellular carcinoma. Biomed Pharmacother 2017;85:348-54.
144. Wang S, Aurora AB, Johnson BA, Qi X, McAnally J, Hill JA, Richardson JA, Bassel-Duby R, Olson EN. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell 2008;15:261-71.
145. Fish JE, Santoro MM, Morton SU, Yu S, Yeh RF, Wythe JD, Ivey KN, Bruneau BG, Stainier DY, Srivastava D. miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell 2008;15:272-84.
146. Yu X, Zheng H, Chan MT V, Wu WK. HULC: an oncogenic long non-coding RNA in human cancer. J Cell Mol Med 2017;21:410-7.
147. Panzitt K, Tschernatsch MMO, Guelly C, Moustafa T, Stradner M, Strohmaier HM, Buck CR, Denk H, Schroeder R, Trauner M, Zatloukal K. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology 2007;132:330-42.
148. Zhao Y, Guo Q, Chen J, Hu J, Wang S, Sun Y. Role of long non-coding RNA HULC in cell proliferation, apoptosis and tumor metastasis of gastric cancer: a clinical and in vitro investigation. Oncol Rep 2014;31:358-64.
149. Hämmerle M, Gutschner T, Uckelmann H, Ozgur S, Fiskin E, Gross M, Skawran B, Geffers R, Longerich T, Breuhahn K, Schirmacher P, Stoecklin G, Diederichs S. Posttranscriptional destabilization of the liver-specific long noncoding RNA HULC by the IGF2 mRNA-binding protein 1 (IGF2BP1). Hepatology 2013;58:1703-12.
150. Pangas SA. Regulation of the ovarian reserve by members of the transforming growth factor beta family. Mol Reprod Dev 2012;79:666-79.
151. Lewis-Wambi JS, Jordan VC. Estrogen regulation of apoptosis: how can one hormone stimulate and inhibit? Breast Cancer Res 2009;11:206.
152. Oesterreich S, Deng W, Jiang S, Cui X, Ivanova M, Schiff R, Kang K, Hadsell DL, Behrens J, Lee AV. Estrogen-mediated down-regulation of E-cadherin in breast cancer cells. Cancer Res 2003;63:5203-8.
153. Lin Z, Reierstad S, Huang CC, Bulun SE. Novel estrogen receptor-α binding sites and estradiol target genes identified by chromatin immunoprecipitation cloning in breast cancer. Cancer Res 2007;67:5017-24.
154. Tan X, Wang S, Yang B, Zhu L, Yin B, Chao T, Zhao J, Yuan J, Qiang B, Peng X. The CREB-miR-9 negative feedback minicircuitry coordinates the migration and proliferation of glioma cells. PLoS One 2012;7:e49570.
155. Schmidt K, Joyce CE, Buquicchio F, Brown A, Ritz J, Distel RJ, Yoon CH, Novina CD. The lncRNA SLNCR1 mediates melanoma invasion through a conserved SRA1-like region. Cell Rep 2016;15:2025-37.
156. Ling H, Spizzo R, Atlasi Y, Nicoloso M, Shimizu M, Redis RS, Nishida N, Gafà R, Song J, Guo Z, Ivan C, Barbarotto E, De Vries I, Zhang X, Ferracin M, Churchman M, van Galen JF, Beverloo BH, Shariati M, Haderk F, Estecio MR, Garcia-Manero G, Patijn GA, Gotley DC, Bhardwaj V, Shureiqi I, Sen S, Multani AS, Welsh J, Yamamoto K, Taniguchi I, Song MA, Gallinger S, Casey G, Thibodeau SN, Le Marchand L, Tiirikainen M, Mani SA, Zhang W, Davuluri RV, Mimori K, Mori M, Sieuwerts AM, Martens JW, Tomlinson I, Negrini M, Berindan-Neagoe I, Foekens JA, Hamilton SR, Lanza G, Kopetz S, Fodde R, Calin GA. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res 2013;23:1446-61.
157. Qiu M, Xu Y, Yang X, Wang J, Hu J, Xu L, Yin R. CCAT2 is a lung adenocarcinoma-specific long non-coding RNA and promotes invasion of non-small cell lung cancer. Tumor Biol 2014;35:5375-80.
158. Yang MH, Hu ZY, Xu C, Xie LY, Wang XY, Chen SY, Li ZG. MALAT1 promotes colorectal cancer cell proliferation/migration/invasion via PRKA kinase anchor protein 9. Biochim Biophys Acta 2015;1852:166-74.
159. Yu Y, Nangia-Makker P, Farhana L, Majumdar APN. A novel mechanism of lncRNA and miRNA interaction: CCAT2 regulates miR-145 expression by suppressing its maturation process in colon cancer cells. Mol Cancer 2017;16:155.
160. Taelman VF, Dobrowolski R, Plouhinec JL, Fuentealba LC, Vorwald PP, Gumper I, Sabatini DD, De Robertis EM. Wnt signaling requires sequestration of glycogen synthase kinase 3 inside multivesicular endosomes. Cell 2010;143:1136-48.
161. Li VS, Ng SS, Boersema PJ, Low TY, Karthaus WR, Gerlach JP, Mohammed S, Heck AJ, Maurice MM, Mahmoudi T, Clevers H. Wnt signaling through Inhibition of β-catenin degradation in an intact Axin1 complex. Cell 2012;149:1245-56.
162. Ling H, Fabbri M, Calin GA. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov 2013;12:847-65.
163. Zhang X, Xu Y, He C, Guo X, Zhang J, He C, Zhang L, Kong M, Chen B, Zhu C. Elevated expression of CCAT2 is associated with poor prognosis in esophageal squamous cell carcinoma. J Surg Oncol 2015;111:834-9.
164. Zhao Z, Wang J, Wang S, Chang H, Zhang T, Qu J. LncRNA CCAT2 promotes tumorigenesis by over-expressed Pokemon in non-small cell lung cancer. Biomed Pharmacother 2017;87:692-7.
165. Cai Y, He J, Zhang D. Long noncoding RNA CCAT2 promotes breast tumor growth by regulating the wnt signaling pathway. Onco Targets Ther 2015;8:2657-64.
166. Redis RS, Vela LE, Lu W, Ferreira de Oliveira J, Ivan C, Rodriguez-Aguayo C, Adamoski D, Pasculli B, Taguchi A, Chen Y, Fernandez AF, Valledor L, Van Roosbroeck K, Chang S, Shah M, Kinnebrew G, Han L, Atlasi Y, Cheung LH, Huang GY, Monroig P, Ramirez MS, Catela Ivkovic T, Van L, Ling H, Gafà R, Kapitanovic S, Lanza G, Bankson JA, Huang P, Lai SY, Bast RC, Rosenblum MG, Radovich M, Ivan M, Bartholomeusz G, Liang H, Fraga MF, Widger WR, Hanash S, Berindan-Neagoe I, Lopez-Berestein G, Ambrosio ALB, Gomes Dias SM, Calin GA. Allele-specific reprogramming of cancer metabolism by the long non-coding RNA CCAT2. Mol Cell 2016;61:520-34.
167. Redis RS, Sieuwerts AM, Look MP, Tudoran O, Ivan C, Spizzo R, Zhang X, de Weerd V, Shimizu M, Ling H, Buiga R, Pop V, Irimie A, Fodde R, Bedrosian I, Martens JW, Foekens JA, Berindan-Neagoe I, Calin GA. CCAT2, a novel long non-coding RNA in breast cancer: expression study and clinical correlations. Oncotarget 2013;4:1748-62.
168. Wang J, Qiu M, Xu Y, Li M, Dong G, Mao Q, Yin R, Xu L. Long noncoding RNA CCAT2 correlates with smoking in esophageal squamous cell carcinoma. Tumour Biol 2015;36:5523-8.
169. Vausort M, Wagner DR, Devaux Y. Long noncoding RNAs in patients with acute myocardial infarction. Circ Res 2014;115:668-77.
170. Ishii N, Ozaki K, Sato H, Mizuno H, Saito S, Takahashi A, Miyamoto Y, Ikegawa S, Kamatani N, Hori M, Saito S, Nakamura Y, Tanaka T. Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction. J Hum Genet 2006;51:1087-99.
171. Rao SQ, Hu HL, Ye N, Shen Y, Xu Q. Genetic variants in long non-coding RNA MIAT contribute to risk of paranoid schizophrenia in a Chinese Han population. Schizophr Res 2014;166:125-30.
172. Hu N, Strobl-Mazzulla PH, Bronner ME. Epigenetic regulation in neural crest development. Dev Biol 2014;396:159-68.
173. Alipoor FJ, Asadi MH, Torkzadeh-Mahani M. Miat lncRNA is overexpressed in breast cancer and its inhibition triggers senescence and G1 arrest in MCF7 cell line. J Cell Biochem 2018; doi: 10.1002/jcb.26678.
174. Broderick JA, Zamore PD. Competitive endogenous RNAs cannot alter microRNA function in vivo. Mol Cell 2014;54:711-3.
175. Taulli R, Loretelli C, Pandolfi PP. From pseudo-ceRNAs to circ-ceRNAs: a tale of cross-talk and competition. Nat Struct Mol Biol 2013;20:541-3.
177. Liu Y, Sánchez-Tilló E, Lu X, Huang L, Clem B, Telang S, Jenson AB, Cuatrecasas M, Chesney J, Postigo A, Dean DC. Sequential inductions of the ZEB1 transcription factor caused by mutation of Rb and then Ras proteins are required for tumor initiation and progression. J Biol Chem 2013;288:11572-80.
178. Shi C, Ren L, Sun C, Yu L, Bian X, Zhou X, Wen Y, Hua D, Zhao S, Luo W, Wang R, Rao C, Wang Q, Yu S. MiR-29a/b/c function as invasion suppressors for gliomas by targeting CDC42 and predict the prognosis of patients. Br J Cancer 2017;117:1036-47.
179. Qiu F, Sun R, Deng N, Guo T, Cao Y, Yu Y, Wang X, Zou B, Zhang S, Jing T, Ling T, Xie J, Zhang Q. MiR-29a/b enhances cell migration and invasion in nasopharyngeal carcinoma progression by regulating SPARC and COL3A1 gene expression. PLoS One 2015;10:e0120969.
180. Gooding AJ, Zhang B, Jahanbani FK, Gilmore HL, Chang JC, Valadkhan S, Schiemann WP. The lncRNA BORG drives breast cancer metastasis and disease recurrence. Sci Rep 2017;7:12698.
181. Addison JB, Koontz C, Fugett JH, Creighton CJ, Chen D, Farrugia MK, Padon RR, Voronkova MA, McLaughlin SL, Livengood RH, Lin CC, Ruppert JM, Pugacheva EN, Ivanov AV. KAP1 promotes proliferation and metastatic progression of breast cancer cells. Cancer Res 2015;75:344-55.
182. Lee YK, Thomas SN, Yang AJ, Ann DK. Doxorubicin down-regulates Krüppel-associated box domain-associated protein 1 sumoylation that relieves its transcription repression on p21WAF1/CIP1 in breast cancer MCF-7 cells. J Biol Chem 2007;282:1595-606.
183. Prensner JR, Chen W, Han S, Iyer MK, Cao Q, Kothari V, Evans JR, Knudsen KE, Paulsen MT, Ljungman M, Lawrence TS, Chinnaiyan AM, Feng FY. The long non-coding RNA PCAT-1 promotes prostate cancer cell proliferation through cMyc. Neoplasia 2014;16:900-8.
184. Prensner JR, Chen W, Iyer MK, Cao Q, Ma T, Han S, Sahu A, Malik R, Wilder-Romans K, Navone N, Logothetis CJ, Araujo JC, Pisters LL, Tewari AK, Canman CE, Knudsen KE, Kitabayashi N, Rubin MA, Demichelis F, Lawrence TS, Chinnaiyan AM, Feng FY. PCAT-1, a long noncoding RNA, regulates BRCA2 and controls homologous recombination in cancer. Cancer Res 2014;74:1651-60.
185. Feng FY, Prensner JR, Chen W, Iyer MK, Cao Q, Wilder-Romans K, Lawrence TS, Chinnaiyan AM. A tissue-specific long noncoding RNA controls homologous recombination in prostate cancer. Int J Radiat Oncol 2012;84:S176.
186. Guo H, Ahmed M, Zhang F, Yao CQ, Li S, Liang Y, Hua J, Soares F, Sun Y, Langstein J, Li Y, Poon C, Bailey SD, Desai K, Fei T, Li Q, Sendorek DH, Fraser M, Prensner JR, Pugh TJ, Pomerantz M, Bristow RG, Lupien M, Feng FY, Boutros PC, Freedman ML, Walsh MJ, He HH. Modulation of long noncoding RNAs by risk SNPs underlying genetic predispositions to prostate cancer. Nat Genet 2016;48:1142-50.
188. Chi SW, Zang JB, Mele A, Darnell RB. Ago HITS-CLIP decodes miRNA-mRNA interaction maps. Nature 2009;460:479-86.
189. Helwak A, Kudla G, Dudnakova T, Tollervey D. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell 2013;153:654-65.
190. Tano K, Mizuno R, Okada T, Rakwal R, Shibato J, Masuo Y, Ijiri K, Akimitsu N. MALAT-1 enhances cell motility of lung adenocarcinoma cells by influencing the expression of motility-related genes. FEBS Lett 2010;584:4575-80.
191. Ji P, Diederichs S, Wang W, Böing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, Thomas M, Berdel WE, Serve H, Müller-Tidow C. MALAT-1, a novel noncoding RNA, and thymosin β4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 2003;22:8031-41.
192. Ying L, Chen Q, Wang Y, Zhou Z, Huang Y, Qiu F. Upregulated MALAT-1 contributes to bladder cancer cell migration by inducing epithelial-to-mesenchymal transition. Mol Biosyst 2012;8:2289-94.
193. Zhang Y, Wang L, Li N, Dai H, Sun J, Cai HF. Downregulation of long non-coding RNA MALAT1 induces tumor progression of human breast cancer through regulating CCND1 expression. Open Life Sci 2016;11:232-6.
194. Liu JH, Chen G, Dang YW, Li CJ, Luo DZ. Expression and prognostic significance of lncRNA MALAT1 in pancreatic cancer tissues. Asian Pac J Cancer Prev 2014;15:2971-7.
195. Zhou Y, Zhang X, Klibanski A. MEG3 noncoding RNA: a tumor suppressor. J Mol Endocrinol 2012;48:R45-53.
196. Xiu YL, Sun KX, Chen X, Chen S, Zhao Y, Guo QG, Zong ZH. Upregulation of the lncRNA Meg3 induces autophagy to inhibit tumorigenesis and progression of epithelial ovarian carcinoma by regulating activity of ATG3. Oncotarget 2017;8:31714-25.
197. Zhou Y, Zhong Y, Wang Y, Zhang X, Batista DL, Gejman R, Ansell PJ, Zhao J, Weng C, Klibanski A. Activation of p53 by MEG3 non-coding RNA. J Biol Chem 2007;282:24731-42.
198. Lu KH, Li W, Liu XH, Sun M, Zhang ML, Wu WQ, Xie WP, Hou YY. Long non-coding RNA MEG3 inhibits NSCLC cells proliferation and induces apoptosis by affecting p53 expression. BMC Cancer 2013;13:461.
199. Peng W, Si S, Zhang Q, Li C, Zhao F, Wang F, Yu J, Ma R. Long non-coding RNA MEG3 functions as a competing endogenous RNA to regulate gastric cancer progression. J Exp Clin Cancer Res 2015;34:79.
200. Li H, An J, Wu M, Zheng Q, Gui X, Li T, Pu H, Lu D. LncRNA HOTAIR promotes human liver cancer stem cell malignant growth through downregulation of SETD2. Oncotarget 2015;6:27847-64.
201. Qiu JJ, Lin YY, Ye LC, Ding JX, Feng WW, Jin HY, Zhang Y, Li Q, Hua KQ. Overexpression of long non-coding RNA HOTAIR predicts poor patient prognosis and promotes tumor metastasis in epithelial ovarian cancer. Gynecol Oncol 2014;134:121-8.
202. Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010;464:1071-6.
203. Tao S, He H, Chen Q. Estradiol induces HOTAIR levels via GPER-mediated miR-148a inhibition in breast cancer. J Transl Med 2015;13:131.
204. Kim K, Jutooru I, Chadalapaka G, Johnson G, Frank J, Burghardt R, Kim S, Safe S. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene 2013;32:1616-25.
205. Hu L, Ye H, Huang G, Luo F, Liu Y, Liu Y, Yang X, Shen J, Liu Q, Zhang J. Long noncoding RNA GAS5 suppresses the migration and invasion of hepatocellular carcinoma cells via miR-21. Tumor Biol 2016;37:2691-702.
206. Zhang Z, Zhu Z, Watabe K, Zhang X, Bai C, Xu M, Wu F, Mo YY. Negative regulation of lncRNA GAS5 by miR-21. Cell Death Differ 2013;20:1558-68.
207. Sun M, Jin FY, Xia R, Kong R, Li JH, Xu TP, Liu YW, Zhang EB, Liu XH, De W. Decreased expression of long noncoding RNA GAS5 indicates a poor prognosis and promotes cell proliferation in gastric cancer. BMC Cancer 2014;14:319.
208. Pickard MR, Williams GT. Regulation of apoptosis by long non-coding RNA GAS5 in breast cancer cells: implications for chemotherapy. Breast Cancer Res Treat 2014;145:359-70.
209. Matouk IJ, DeGroot N, Mezan S, Ayesh S, Abu-lail R, Hochberg A, Galun E. The H19 non-coding RNA is essential for human tumor growth. PLoS One 2007;2:e845.
210. Matouk IJ, Halle D, Raveh E, Gilon M, Sorin V, Hochberg A. The role of the oncofetal H19 lncRNA in tumor metastasis: orchestrating the EMT-MET decision. Oncotarget 2016;7:3748-65.
211. Matouk IJ, Raveh E, Abu-lail R, Mezan S, Gilon M, Gershtain E, Birman T, Gallula J, Schneider T, Barkali M, Richler C, Fellig Y, Sorin V, Hubert A, Hochberg A, Czerniak A. Oncofetal H19 RNA promotes tumor metastasis. Biochim Biophys Actam 2014;1843:1414-26.
212. Ma C, Nong K, Zhu H, Wang W, Huang X, Yuan Z, Ai K. H19 promotes pancreatic cancer metastasis by derepressing let-7's suppression on its target HMGA2-mediated EMT. Tumor Biol 2014;35:9163-9.
213. Yang F, Bi J, Xue X, Zheng L, Zhi K, Hua J, Fang G. Up-regulated long non-coding RNA H19 contributes to proliferation of gastric cancer cells. FEBS J 2012;279:3159-65.
214. Matouk IJ, Abbasi I, Hochberg A, Galun E, Dweik H, Akkawi M. Highly upregulated in liver cancer noncoding RNA is overexpressed in hepatic colorectal metastasis. Eur J Gastroenterol Hepatol 2009;21:688-92.
215. Sun M, Liu XH, Lu KH, Nie FQ, Xia R, Kong R, Yang JS, Xu TP, Liu YW, Zou YF, Lu BB, Yin R, Zhang EB, Xu L, De W, Wang ZX. EZH2-mediated epigenetic suppression of long noncoding RNA SPRY4-IT1 promotes NSCLC cell proliferation and metastasis by affecting the epithelial-mesenchymal transition. Cell Death Dis 2014;5:e1298.
216. Fang D, Yang H, Lin J, Teng Y, Jiang Y, Chen J, Li Y. 17β-Estradiol regulates cell proliferation, colony formation, migration, invasion and promotes apoptosis by upregulating miR-9 and thus degrades MALAT-1 in osteosarcoma cell MG-63 in an estrogen receptor-independent manner. Biochem Biophys Res Commun 2015;457:500-6.
217. Jin C, Yan B, Lu Q, Lin Y, Ma L. Reciprocal regulation of Hsa-miR-1 and long noncoding RNA MALAT1 promotes triple-negative breast cancer development. Tumor Biol 2016;37:7383-94.
218. Tang Y, Jin X, Xiang Y, Chen Y, Shen CX, Zhang YC, Li YG. The lncRNA MALAT1 protects the endothelium against ox-LDL-induced dysfunction via upregulating the expression of the miR-22-3p target genes CXCR2 and AKT. FEBS Lett 2015;589:3189-96.
219. Huang X, Lu S. MicroR-545 mediates colorectal cancer cells proliferation through up-regulating epidermal growth factor receptor expression in HOTAIR long non-coding RNA dependent. Mol Cell Biochem 2017;431:45-54.
220. Xu F, Zhang J. Long non-coding RNA HOTAIR functions as miRNA sponge to promote the epithelial to mesenchymal transition in esophageal cancer. Biomed Pharmacother 2017;90:888-96.
221. Li JT, Wang LF, Zhao YL, Yang T, Li W, Zhao J, Yu F, Wang L, Meng YL, Liu NN, Zhu XS, Gao CF, Jia LT, Yang AG. Nuclear factor of activated T cells 5 maintained by Hotair suppression of miR-568 upregulates S100 calcium binding protein A4 to promote breast cancer metastasis. Breast Cancer Res 2014;16:454.
222. Lei K, Liang X, Gao Y, Xu B, Xu Y, Li Y, Tao Y, Shi W, Liu J. Lnc-ATB contributes to gastric cancer growth through a MiR-141-3p/TGFβ2 feedback loop. Biochem Biophys Res Commun 2017;484:514-21.
223. Zhou X, Ye F, Yin C, Zhuang Y, Yue G, Zhang G. The interaction between MiR-141 and lncRNA-H19 in regulating cell proliferation and migration in gastric cancer. Cell Physiol Biochem 2015;36:1440-52.
224. Zhu M, Chen Q, Liu X, Sun Q, Zhao X, Deng R, Wang Y, Huang J, Xu M, Yan J, Yu J. LncRNA H19/miR-675 axis represses prostate cancer metastasis by targeting TGFBI. FEBS J 2014;281:3766-75.
225. Keniry A, Oxley D, Monnier P, Kyba M, Dandolo L, Smits G, Reik W. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat Cell Biol 2012;14:659-65.
226. Lu Y, Li Y, Chai X, Kang Q, Zhao P, Xiong J, Wang J. Long noncoding RNA HULC promotes cell proliferation by regulating PI3K/AKT signaling pathway in chronic myeloid leukemia. Gene 2017;607:41-6.
228. Hou P, Zhao Y, Li Z, Yao R, Ma M, Gao Y, Zhao L, Zhang Y, Huang B, Lu J. LincRNA-ROR induces epithelial-to-mesenchymal transition and contributes to breast cancer tumorigenesis and metastasis. Cell Death Dis 2014;5:e1287.
Cite This Article
Export citation file: BibTeX | RIS
OAE Style
Parsons C, Tayoun AM, Benado BD, Ragusa G, Dorvil RF, Rourke EA, Connor KO, Reed IG, Alexander A, Willetts L, Habibian M, Adams BD. The role of long noncoding RNAs in cancer metastasis. J Cancer Metastasis Treat 2018;4:19. http://dx.doi.org/10.20517/2394-4722.2018.11
AMA Style
Parsons C, Tayoun AM, Benado BD, Ragusa G, Dorvil RF, Rourke EA, Connor KO, Reed IG, Alexander A, Willetts L, Habibian M, Adams BD. The role of long noncoding RNAs in cancer metastasis. Journal of Cancer Metastasis and Treatment. 2018; 4: 19. http://dx.doi.org/10.20517/2394-4722.2018.11
Chicago/Turabian Style
Parsons, Christine, Alexis M. Tayoun, Benjamin D. Benado, Gabriel Ragusa, Rose F. Dorvil, Elizabeth A. Rourke, Kaitlyn O’ Connor, Irene G. Reed, Angela Alexander, Lian Willetts, Maryam Habibian, Brian D. Adams. 2018. "The role of long noncoding RNAs in cancer metastasis" Journal of Cancer Metastasis and Treatment. 4: 19. http://dx.doi.org/10.20517/2394-4722.2018.11
ACS Style
Parsons, C.; Tayoun AM.; Benado BD.; Ragusa G.; Dorvil RF.; Rourke EA.; Connor KO.; Reed IG.; Alexander A.; Willetts L.; Habibian M.; Adams BD. The role of long noncoding RNAs in cancer metastasis. J. Cancer. Metastasis. Treat. 2018, 4, 19. http://dx.doi.org/10.20517/2394-4722.2018.11
About This Article
Copyright
Data & Comments
Data
 Cite This Article 13 clicks
Cite This Article 13 clicks



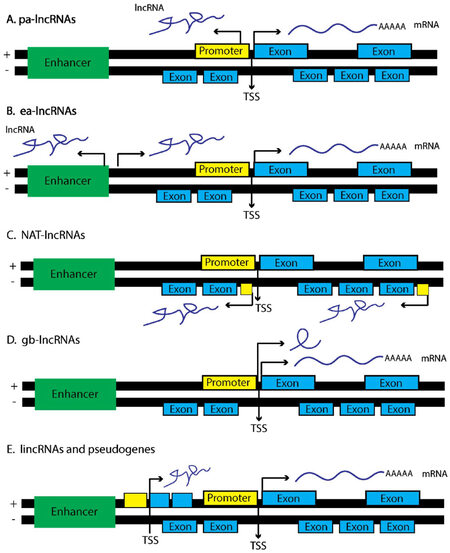
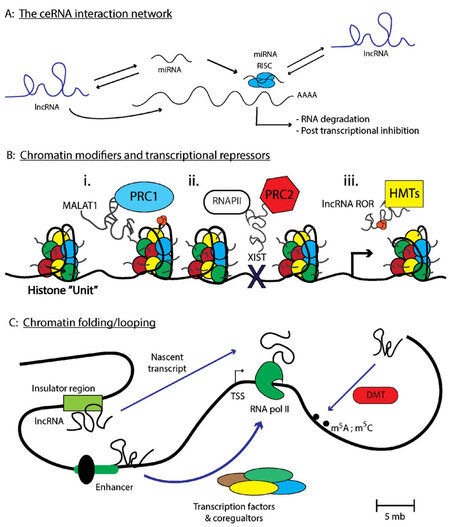









Comments
Comments must be written in English. Spam, offensive content, impersonation, and private information will not be permitted. If any comment is reported and identified as inappropriate content by OAE staff, the comment will be removed without notice. If you have any queries or need any help, please contact us at support@oaepublish.com.