Contribution of alternative splicing to breast cancer metastasis
Abstract
Alternative splicing is a major contributor to transcriptome and proteome diversity in eukaryotes. Comparing to normal samples, about 30% more alternative splicing events were recently identified in 32 cancer types included in The Cancer Genome Atlas database. Some alternative splicing isoforms and their encoded proteins contribute to specific cancer hallmarks. In this review, we will discuss recent progress regarding the contributions of alternative splicing to breast cancer metastasis. We plan to dissect the role of MTDH, CD44 and their interaction with other mRNA splicing factors. We believe an in-depth understanding of the mechanism underlying the contribution of splicing to breast cancer metastasis will provide novel strategies to the management of breast cancer.
Keywords
Introduction
Breast cancer is the most common type of cancer among women. In despite of emerging new treatments such as PARP inhibition and immune checkpoint blockade, it remains a major challenge[1,2] and is the primary cause of cancer mortality in women. In the majority of cases, the death from breast cancer is not due to the primary tumor per se, but rather the result of metastasis to other organs in the body[3]. Metastasis is a multi-step process involving stromal invasion, cell migration, intravasation, anoikis resistance, extravasation and subsequent implantation and proliferation in sites other than the primary location[4]. Although we have gained ample knowledge in this cellular process, an in-depth understanding at the molecular level remains to be deciphered.
Alternative splicing might be such a molecular mechanism that contributes to metastasis. It is a process whereby multiple functionally distinct transcripts are encoded from a single gene by the selective removal or retention of exons and/or introns from the maturing RNA. This process is highly regulated, involving trans-acting splicing factors and cis-acting regulatory motifs and so is susceptible to hereditary and somatic mutations. Alternative splicing is common in many eukaryote lineages. Using deep transcriptome sequencing of the human genome, over 95% of multi-exon genes were found capable of producing at least one alternatively spliced isoform[5]. Many single-gene studies have also characterized the role of alternative splicing in various cellular processes. Disruption or dysregulation of alternative splicing has also been associated with pathological states[6,7]. Maguire et al.[8] demonstrated that spliceosomal mutations occur in a mutually exclusive manner in breast cancer and that distinct components of the spliceosome are targeted by somatic mutations in different types of breast cancer. The exact splicing pattern associated with a particular breast cancer type or stage still requires a broad characterization through molecular analysis of splicing isoforms in different patients. However, existing evidence strongly supports a pivotal role of alternative splicing in breast cancer biology and innovative tools are under development to use splicing events for diagnostic and therapeutic purposes[9]. Shapiro et al.[10] observed an epithelial-mesenchymal transition (EMT)-associated global change in alternative splicing of a number of genes that are involved in functions crucial for EMT progression, such as cell adhesion, cell motility, and cytoskeletal remodeling. Several of the splicing changes discovered in vitro were also found to occur in a panel of breast cancer cell lines and in vivo in primary human breast cancer samples. Dysregulation of alternative splicing has been increasingly recognized in cancer-related pathways. It is thus critical to investigate the functional significance of splicing regulation in the context of cancer. This review will discuss some recent progresses about the alternative splicing regulators such as CD44, heterogeneous nuclear ribonucleoprotein M (hnRNPM), SND1 and MTDH etc. in breast cancer metastasis.
Splicing variants may enhance EMT and metastasis in breast cancer
Dorman et al.[12] reported splicing defects in large-scale breast cancer sequencing studies. Nine hundred and eighty-eight splicing variants including exon skipping, leaky or cryptic splicing from 5,206 putative mutations were confirmed for splicing mutations in 442 Breast Cancer patients from The Cancer Genome Atlas dataset. These splicing variants were significantly increased in patients with lymph node metastasis, but not in lymph node-negative tumors. Silipo et al.[13] reported that the expression profile changes of splicing factors including serine/arginine-rich splicing factor 1 (SRSF1), SRSF2, SRSF3, SRSF5, SRSF6 and SRSF10; the heterogeneous nuclear ribonucleoproteins (hnRNPs) including hnRNP A2/B1, hnRNPI, hnRNPA1 and hnRNP K; as well as eight RNA-binding proteins including HuR, Sam68, BRM5, FOX2, YB-1, PRMT6, SPF45 and PELP1 in breast cancer cells compared with normal cells, which are strongly associated with the alternative splicing pattern of many cancer-related genes despite the absence of mutations in genomic DNA. Inoue et al.[14] reported aberrant splicing of CD44 gene in breast cancer, which promotes metastasis. The status of splicing factors and other splicing-related proteins in breast cancer are important to provide insights into the mechanisms that lead to breast cancer metastasis[15]. The correlation analysis of somatic variants with alternative splicing events confirmed known trans- associations with variants in SF3B1 and U2AF1, and additional trans-acting variants (e.g., TADA1, PPP2R1A). Tumors have up to 30% more alternative splicing events than normal samples. Many tumors have thousands of alternative splicing events that are not detectable in normal samples. On average, 930 exon-exon junctions (‘‘neojunctions’’) were identified in tumors not typically found in normal tissue included in the Genotype-Tissue Expression (GTEx) project[15]. CD44 is a cell surface protein with various isoforms that involves in motility, cell survival and proliferation and the formation of tumor microenvironment. Alternative splicing can produce various isoforms of CD44 with properties at different specific tissue[16,17]. The RNA-binding protein hnRNPM was found to promote breast cancer metastasis by activating the switch of alternative splicing during EMT.
Hnrnpm increases CD44 alternative splicing to enhance breast cancer metastasis
CD44 was identified as a key downstream target of hnRNPM by genome-wide deep sequencing analysis. hnRNPM is associated with increased standard form of CD44 (or CD44 standard, CD44s) in aggressive breast cancer patient specimens. Overexpressed hnRNPM competes with epithelial splicing regulatory protein 1 (ESRP1), and binds to the same cis-regulatory RNA elements of CD44 for the precisely control CD44s splice isoform switching during EMT[18]. ESRP1 is a splicing regulator to promote an epithelial splicing program and hnRNPM is a mesenchymal splicing regulator. Harvey et al.[19] reported that hnRNPM and ESRP1 co-regulate a set of cassette exon events in EMT genes associated with cell migration and cytoskeletal reorganization. Competitive binding to these cis-elements by hnRNPM and ESRP1 to antagonize alternative splicing was proposed. The expression of hnRNPM is closely correlated with invasion and metastasis of tumor cells. hnRNPM expression was upregulated in breast cancer tissues. HnRNPM and CD44s expression are positively correlated in breast cancer tissues. Cancer stem cells marker ALDH1+ was found positively associated with overexpression of CD44s and hnRNPM. High hnRNPM is associated with higher levels of CD44s, shorter overall survival and higher rates of lymph node metastases in breast cancer patients[20].
MORC2-mutant M276I promotes an hnrnpm-mediated CD44 alternative splicing to enhance breast cancer metastasis
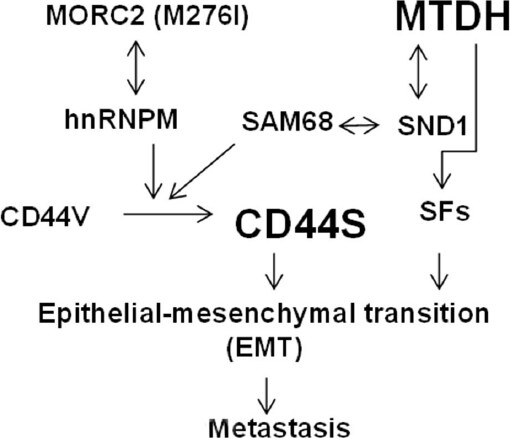
A cancer-associated Microrchidia family CW-type zinc finger 2 (MORC2) (M276I) mutant was reported to promote metastatic ability of TNBC cancer cells by enhancing interaction with hnRNPM and splicing switch of CD44 from the epithelial isoform (CD44v) to the mesenchymal isoform (CD44s) [Figure 1]. Expression of mutant MORC2 in TNBC cells increased cell migration, invasion, and lung metastasis. The M276I mutation enhanced binding of MORC2 to hnRNPM, a component of the spliceosome machinery. This interaction promoted an hnRNPM-mediated splicing switch of CD44 from CD44v) to CD44s, ultimately driving EMT. Knockdown of hnRNPM reduced the binding of mutant MORC2 to CD44 pre-mRNA and reversed the mutant MORC2-induced CD44 splicing switch and EMT. As a consequence, the migratory, invasive, and lung metastatic potential of mutant MORC2-expressing cells was impaired[21].
TDP43, CPEB2A/B and ESRP1/RBFOX2 in breast cancer metastasis
Ke et al.[22] reported that the loss of TDP43 (TAR DNA-binding protein 43), an important splicing regulator involved in the SRSF3 regulated unique splicing of downstream gene PAR3, promotes metastasis in TNBC. Highly expressed TDP43 is correlated with poor prognosis in TNBC. Knockdown of TDP43 inhibits SRSF3 and PAR3 mediated metastasis. Two CPEB2 splicing isoforms with or without exon 4 was reported to mediate opposing effects on cancer-related phenotypes. The CPEB2A isoform, which is produced by exclusion of exon 4 from the mature CPEB2 mRNA, inhibited tumor growth. The CPEB2B splicing isoform with the inclusion of exon 4 into the mature CPEB2 mRNA was overexpressed in aggressive forms of human breast cancer and enhanced cancer metastasis was observed[23]. CPEB2A/B promotes the translation of two critical downstream proteins TWIST1 and HIF1a in the hypoxia/EMT pathway[23]. Splicing factor ratio might be an index of EMT and tumor aggressiveness in breast cancer. In fact, the association of low ESRP1/RBFOX2 ratio with high risk of metastasis in early breast cancer was speculated to be a new early prognostic marker of breast cancer metastasis[24].
RNA binding protein RBM47 inhibits breast cancer metastasis by regulating splicing
RNA binding motif protein 47 (RBM47) was identified as a suppressor of breast cancer metastasis through analysis of clinical breast cancer gene expression datasets, cell line models, and mutation data. Transcriptome-wide HITS-CLIP analysis revealed widespread mRNAs associated with RBM47 by binding to their introns and 3’UTRs. The dickkopf WNT signaling pathway inhibitor 1 (DKK1) is one of downstream mRNAs of RBM47. RBM47 inhibits breast cancer metastasis by increasing stability of the Wnt antagonist DKK1[25].
Non-coding RNA rest-003 promotes breast cancer metastasis
Non-coding RNAs (ncRNAs) RE1-silencing transcription factor (REST)-003 was reported to promote breast cancer metastasis. REST-003 is cRNAs derived from the first exon of an alternatively spliced REST transcript processed by serine/arginine repeat-related protein SRRM3. REST is a transcription factor to regulate expression of genes important for neuronal development. Interestingly, SRRM3 expression is repressed by REST[26].
MTDH promotes breast cancer metastasis partially by regulating alternative splicing
High metadherin gene expression was highly correlated with breast cancer metastasis[27,28]. MTDH was identified to bind to the vasculature of the lung by phage display screening cDNAs from metastatic breast carcinoma[27]. Experimental metastasis can be inhibited by metadherin specific antibody or siRNAs. Hu et al reported that MTDH drives breast cancer metastasis to the lungs by increasing adhesion to the walls of blood vessels[28]. Amplification of a minimal 2.9 Mb piece of chromosome 8q22 was identified in poor-prognosis breast cancers by ACE (analysis of CNAs by expression data) and fluorescence in situ hybridization (FISH) analysis. Only the enforced expression of MTDH in this amplified 8q22 region was identified to increase lung seeding after tail vein injection of the mildly metastatic breast cancer cell line MDA-MB-231[28]. Interaction of MTDH with Staphylococcal nuclease domain-containing 1 (SND1) was independently identified by mass spectrometry (MS) by three labs[29-31]. Overexpression of MTDH and SND1 in primary tumors is strongly associated with reduced metastasis-free survival in multiple large-scale datasets of breast cancer patients[32,33]. SND1 acts as a novel alternative splicing regulator by interacting with SAM68 to regulate exon v5 inclusion in the CD44 mRNA splicing that promotes cancer metastasis [Figure 1][34-36]. Several other splicing regulators including hnRNPA0, hnRNPA2B1, hnRNPF, hnRNPA3 isoform were also identified by MS in MTDH pull-down assay[29]. High-throughput sequencing of RNA isolated by Cross-linking immunoprecipitation (HITS-CLIP) and Photoactivatable Ribonucleoside-Enhanced Cross-linking and Immunoprecipitation (PAR-CLIP) were recently developed methods to study RNA-protein interactions[37,38]. As shown in Table 1, mRNAs encoding for mRNA splicing regulators were identified in MTDH RNA interactome for multiple times at different sites by MTDH antibody specific PAR-CLIP and 12 splicing factors were confirmed by MTDH HITS-CLIP[39]. Therefore, MTDH may promote breast cancer metastasis by regulating mRNA splicing through interacting with mRNAs or proteins of splicing factors.
List of mRNAs encoding for mRNA splicing factors identified by MTDH PAR-CLIP or HITS-CLIP
| Ref Seq# | Gene | #PAR-CLIP | Ref Seq# | Gene | #PAR-CLIP |
|---|---|---|---|---|---|
| NM_006924 | SRSF1 | 16 | NM_004698 | PRPF3 | 6 |
| NM_003016 | SRSF2 | 14 | NM_014502 | PRPF19 | 11 |
| NM_003017 | SRSF3* | 7 | NM_015629 | PRPF31 | 3 |
| NM_005626 | SRSF4 | 8 | NM_006109 | PRMT5 | 7 |
| NM_001039465 | SRSF5 | 9 | NM_014706 | SART3 | 8 |
| NM_006275 | SRSF6 | 13 | NM_018047 | RBM22 | 5 |
| NM_001031684 | SRSF7 | 5 | NM_012321 | LSM4 | 5 |
| NM_003769 | SRSF9 | 7 | NM_000344 | SMN1 | 1 |
| NM_054016 | SRSF10 | 5 | NM_015721 | GEMIN4 | 11 |
| NM_001190987 | SRSF11 | 10 | NM_015465 | GEMIN5 | 4 |
| NM_003090 | SNRPA1 | 5 | NM_024707 | GEMIN7 | 3 |
| NM_198216 | SNRPB* | 4 | NM_024707 | HNRNPA1* | 7 |
| NM_003093 | SNRPC | 5 | NM_031157 | HNRNPA2B1* | 12 |
| NM_006938 | SNRPD1 | 3 | NM_031243 | HNRNPA3* | 29 |
| NM_004597 | SNRPD2* | 5 | NM_194247 | HNRNPH3* | 10 |
| NM_004175 | SNRPD3 | 11 | NM_012207 | HNRNPK* | 10 |
| NM_003094 | SNRPE | 3 | NM_002140 | HNRNPL* | 6 |
| NM_003095 | SNRPF | 2 | NM_001005335 | HNRPU* | 17 |
| NM_003096 | SNRPG | 3 | NM_004501 | HNRNPUL2* | 14 |
| NM_022805 | SNRPN | 4 | NM_001079559 | PCBP1* | 10 |
Conclusion
Increased expression of mRNAs alternative splicing isoforms derived from alteration of splicing factors and MTDH expression could promote EMT and breast cancer metastasis, which provides new targets for breast cancer therapy.
Declarations
Authors’ contributionsConception and elaboration of the work: Meng X, Yang S, Zhang J, Yu H
Provided administrative, technical, and material support: Meng X, Yang S, Zhang J, Yu H
Final approval of the version: Meng X, Yang S, Zhang J, Yu H
Availability of data and materialsNot applicable.
Financial support and sponsorshipThe studies were supported from NIH (RO1CA184101) to Meng X.
Conflicts of interestAll authors declared that there are no conflicts of interest.
Ethical approval and consent to participateNot applicable.
Consent for publicationNot applicable.
Copyright© The Author(s) 2019.
REFERENCES
1. Mansoori B, Mohammadi A, Ghasabi M, Shirjang S, Dehghan R, et al. miR-142-3p as tumor suppressor miRNA in the regulation of tumorigenicity, invasion and migration of human breast cancer by targeting Bach-1 expression. J Cell Physiol 2019;234:9816-25.
2. da Costa AM, Hashim D, Fregnani JHTG, Weiderpass E. Overall survival and time trends in breast and cervical cancer incidence and mortality in the Regional Health District (RHD) of Barretos, São Paulo, Brazil. BMC Cancer 2018;18:1079.
3. Scully OJ, Bay BH, Yip G, Yu Y. Breast cancer metastasis. Cancer Genomics Proteomics 2012;9:311-20.
4. Saharat K, Lirdprapamongkol K, Chokchaichamnankit D, Srisomsap C, Svasti J, et al. Tumor susceptibility gene 101 mediates anoikis resistance of metastatic thyroid cancer cells. Cancer Genomics Proteomics 2018;15:473-83.
5. Bush SJ, Chen L, Tovar-Corona JM, Urrutia AO. Alternative splicing and the evolution of phenotypic novelty. Philos Trans R Soc Lond B Biol Sci 2017;372:20150474.
6. Cieply B, Carstens RP. Functional roles of alternative splicing factors in human disease. Wiley Interdiscip Rev RNA 2015;6:311-26.
8. Maguire SL, Leonidou A, Wai P, Marchiò C, Ng CK, et al. SF3B1 mutations constitute a novel therapeutic target in breast cancer. J Pathol 2015;235:571-80.
9. Martínez-Montiel N, Anaya-Ruiz M, Pérez-Santos M, Martínez-Contreras RD. Alternative splicing in breast cancer and the potential development of therapeutic tools. Genes (Basel) 2017;8:217.
10. Shapiro IM, Cheng AW, Flytzanis NC, Balsamo M, Condeelis JS, et al. An EMT-driven alternative splicing program occurs in human breast cancer and modulates cellular phenotype. PLoS Genet 2011;7:e1002218.
11. Xu Y, Gao XD, Lee JH, Huang H, Tan H, et al. Cell type-restricted activity of hnRNPM promotes breast cancer metastasis via regulating alternative splicing. Genes Dev 2014;28:1191-203.
12. Dorman SN, Viner C, Rogan PK. Splicing mutation analysis reveals previously unrecognized pathways in lymph node-invasive breast cancer. Sci Rep 2014;4:7063.
13. Silipo M, Gautrey H, Tyson-Capper A. Deregulation of splicing factors and breast cancer development. J Mol Cell Biol 2015;7:388-401.
14. Inoue K, Fry EA. Aberrant splicing of estrogen receptor, HER2, and CD44 genes in breast cancer. Genet Epigenet 2015;7:19-32.
15. Kahles A, Lehmann KV, Toussaint NC, Hüser M, Stark SG, et al. Comprehensive analysis of alternative splicing across tumors from 8,705 patients. Cancer Cell 2018;34:211-24.
16. Ishimoto T, Nagano O, Yae T, Tamada M, Motohara T, et al. CD44 variant regulates redox status in cancer cells by stabilizing the xCT subunit of system xc(-) and thereby promotes tumor growth. Cancer Cell 2011;19:387-400.
17. Prochazka L, Tesarik R, Turanek J. Regulation of alternative splicing of CD44 in cancer. Cell Signal 2014;26:2234-9.
18. Xu Y, Gao XD, Lee JH, Huang H, Tan H, et al. Cell type-restricted activity of hnRNPM promotes breast cancer metastasis via regulating alternative splicing. Genes Dev 2014;11:1191-203.
19. Harvey SE, Xu Y, Lin X, Gao XD, Qiu Y, et al. Coregulation of alternative splicing by hnRNPM and ESRP1 during EMT. RNA 2018;24:1326-38.
20. Sun H, Liu T, Zhu D, Dong X, Liu F, et al. HnRNPM and CD44s expression affects tumor aggressiveness and predicts poor prognosis in breast cancer with axillary lymph node metastases. Genes Chromosomes Cancer 2017;56:598-607.
21. Zhang FL, Cao JL, Xie HY, Sun R, Yang LF, et al. Cancer-associated MORC2-mutant M276I regulates an hnRNPM-mediated CD44 splicing switch to promote invasion and metastasis in triple-negative breast cancer. Cancer Res 2018;20:5780-92.
22. Ke H, Zhao L, Zhang H, Feng X, Xu H, et al. Loss of TDP43 inhibits progression of triple-negative breast cancer in coordination with SRSF3. Proc Natl Acad Sci U S A 2018;115:E3426-35.
23. DeLigio JT, Lin G, Chalfant CE, Park MA. Splice variants of cytosolic polyadenylation element-binding protein 2 (CPEB2) differentially regulate pathways linked to cancer metastasis. J Biol Chem 2017;292:17909-18.
24. Fici P, Gallerani G, Morel AP, Mercatali L, Ibrahim T, et al. Splicing factor ratio as an index of epithelial-mesenchymal transition and tumor aggressiveness in breast cancer. Oncotarget 2017;8:2423-36.
25. Vanharanta S, Marney CB, Shu W, Valiente M, Zou Y, et al. Loss of the multifunctional RNA-binding protein RBM47 as a source of selectable metastatic traits in breast cancer. Elife 2014;3:1-24.
26. Lee NS, Evgrafov OV, Souaiaia T, Bonyad A, Herstein J, et al. Non-coding RNAs derived from an alternatively spliced REST transcript (REST-003) regulate breast cancer invasiveness. Sci Rep 2015;5:11207.
27. Brown DM, Ruoslahti E. Metadherin, a cell surface protein in breast tumors that mediates lung metastasis. Cancer Cell 2004;5:365-74.
28. Hu G, Chong RA, Yang Q, Wei Y, Blanco MA, et al. MTDH activation by 8q22 genomic gain promotes chemoresistance and metastasis of poor-prognosis breast cancer. Cancer Cell 2009;15:9-20.
29. Meng X, Zhu D, Yang S, Wang X, Xiong Z, et al. Cytoplasmic Metadherin (MTDH) provides survival advantage under conditions of stress by acting as RNA-binding protein. J Biol Chem 2012;287:4485-91.
30. Blanco MA, Alečković M, Hua Y, Li T, Wei Y, et al. Identification of staphylococcal nuclease domain-containing 1 (SND1) as a Metadherin-interacting protein with metastasis-promoting functions. J Biol Chem 2011;286:19982-92.
31. Yoo BK, Santhekadur PK, Gredler R, Chen D, Emdad L, et al. Increased RNA-induced Silencing complex (RISC) activity contributes to hepatocellular carcinoma. Hepatology 2011;53:1538-48.
32. Meng X, Thiel KW, Leslie KK. Drug resistance mediated by AEG-1/MTDH/LYRIC. Adv Cancer Res 2013;120:135-57.
33. Wan L, Lu X, Yuan S, Wei Y, Guo F, et al. MTDH-SND1 interaction is crucial for expansion and activity of tumor-initiating cells in diverse oncogene- and carcinogen-induced mammary tumors. Cancer Cell 2014;26:92-105.
34. Cappellari M, Bielli P, Paronetto MP, Ciccosanti F, Fimia GM, et al. The transcriptional co-activator SND1 is a novel regulator of alternative splicing in prostate cancer cells. Oncogene 2014;33:3794-802.
35. Gao X, Zhao X, Zhu Y, He J, Shao J, et al. Tudor staphylococcal nuclease (Tudor-SN) participates in small ribonucleoprotein (snRNP) assembly via interacting with symmetrically dimethylated Sm proteins. J Biol Chem 2012;287:18130-41.
36. Yang J, Välineva T, Hong J, Bu T, Yao Z, et al. Transcriptional co-activator protein p100 interacts with snRNP proteins and facilitates the assembly of the spliceosome. Nucleic Acids Res 2007;35:4485-94.
37. Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 2009;460:479-86.
38. Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 2010;141:129-41.
Cite This Article
Export citation file: BibTeX | RIS
OAE Style
Meng X, Yang S, Zhang J, Yu H. Contribution of alternative splicing to breast cancer metastasis. J Cancer Metastasis Treat 2019;5:21. http://dx.doi.org/10.20517/2394-4722.2018.96
AMA Style
Meng X, Yang S, Zhang J, Yu H. Contribution of alternative splicing to breast cancer metastasis. Journal of Cancer Metastasis and Treatment. 2019; 5: 21. http://dx.doi.org/10.20517/2394-4722.2018.96
Chicago/Turabian Style
Meng, Xiangbing, Shujie Yang, Jun Zhang, Huimin Yu. 2019. "Contribution of alternative splicing to breast cancer metastasis" Journal of Cancer Metastasis and Treatment. 5: 21. http://dx.doi.org/10.20517/2394-4722.2018.96
ACS Style
Meng, X.; Yang S.; Zhang J.; Yu H. Contribution of alternative splicing to breast cancer metastasis. J. Cancer. Metastasis. Treat. 2019, 5, 21. http://dx.doi.org/10.20517/2394-4722.2018.96
About This Article
Special Issue
Copyright
Data & Comments
Data
 Cite This Article 33 clicks
Cite This Article 33 clicks












Comments
Comments must be written in English. Spam, offensive content, impersonation, and private information will not be permitted. If any comment is reported and identified as inappropriate content by OAE staff, the comment will be removed without notice. If you have any queries or need any help, please contact us at support@oaepublish.com.