Liquid biopsy in endometrial cancer
Abstract
Liquid biopsy (LB) is an emerging tool for the evaluation of relapse in several cancers and nowadays is used in lung cancer for primary detection and molecular characterization when tumoral tissue is not available. It can represent an innovative biospecimen for the screening, diagnosis, and monitoring of all types of cancer and for monitoring of therapeutic efficacy. LB includes several biofluids such as blood, urine, peritoneal fluid/lavage, and analytes (circulating tumor cells, circulating tumor DNA, long noncoding RNA, microRNA, vesicles, mRNA, and protein) that can play different roles in diagnosis, prognosis, and patient management as well as in the improvement of the knowledge of cancer evolution. Endometrial cancer (EC) is a tumor usually detected at low stage with a good prognosis, but few low risk cases, unexpectedly, can evolve to bad prognosis. Up to now, no molecular target exists to treat advanced stage or to define the evolution of low stage EC. This review focuses on how the LB may help in the management and characterization of patients affected by EC.
Keywords
Introduction
The current use of traditional biopsy in the management of cancer has several limitations in the developing era of precision medicine, with cancer treatment mainly due to the progression of cancer and the onset of resistance to therapies. Sometimes tissue biopsy cannot reflect the heterogeneity of the primary tumor and the revolution of tumor due to the natural course of the disease and under the pressure of treatment. Moreover, traditional biopsy is invasive, it is neither feasible nor practical to perform serial biopsies to guide treatment in real time, and, depending on the site of tumor, biopsy cannot be performed.
Liquid biopsy (LB) has the potential to overcome many of the limitations of traditional biopsy since it is highly tailored and minimally invasive, can be repeated several times, is a cost-effective method, and can be used to screen and monitor response to treatment and to identify the clones involved in the relapse, metastasis, or resistance to treatment. However, many challenges still need to be overcome before LB becomes a reliable and widely available option[1].
LB is represented by fluids that can be collected from the body such as blood, urine, cerebrospinal fluid, lavages, peritoneal fluids, and saliva[2], and the related biomarkers are, consequently, circulating tumor markers such as circulating tumor cells (CTCs), nucleic acids, proteins, and metabolites deriving from tumors.
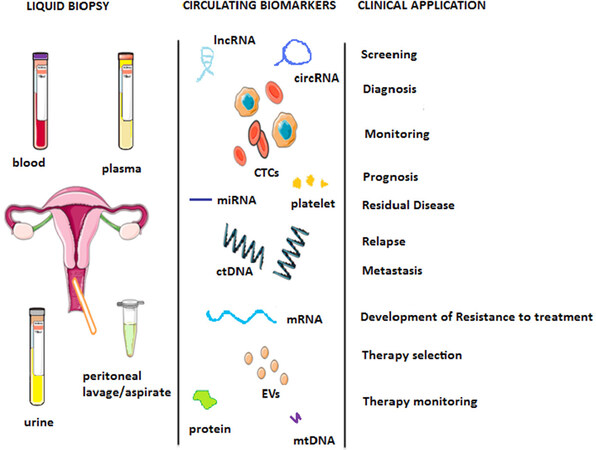
LB usually refers to blood or blood compounds, and the principal biospecimens analyzed are circulating tumor DNA (ctDNA), CTCs, and proteins. Proteins were the first molecules analyzed within blood and other body fluids before the invention of LB [e.g., CA125 in peripheral blood (PB) for ovarian cancer]. Nowadays, the target biomolecules are increasing and include microRNA (miRNA), lncRNA, miRNAs, vesicles, and platelets [Figure 1].
Figure 1. Schematic overview of LB biospecimen, biomarkers, and their clinical applications in EC. Modified from Muinelo-Romay et al.[24]. LB: liquid biopsy; EC: endometrial cancer
Blood circulating biomarkers are currently used in research and have been suggested for tumor characterization to set a personalized target therapy [e.g., epidermal growth factor receptor (EGFR) mutation profile in lung carcinoma] in clinical practice. Despite the great development of droplet-based digital PCR (ddPCR) and the various optimizations of Next Generation Sequencing technologies (NGS), for both CTCs and ctDNA assays, there is no standardization starting from the pre-analytical phase through the analytical procedures to platform/technological methods[3-5].
Endometrial cancer (EC) is the most frequent gynecological malignant disease affecting the inner lining of the uterus[6]. The incidence has increased between 1980 and 2010, due to an increment in average age and obesity[7]. Eighty percent of this type of cancer is diagnosed in an early stage [by International Federation of Gynecology and Obstetrics (FIGO) 2009 classification: stage IA], with five-year survival rates above 80%. EC is classified into two main histological types: Type I, which is more frequent, and Type II, comprising several subtypes such as serous and undifferentiated carcinomas[8,9]. In 2013, the Cancer Genome Atlas Research Network (TCGA) suggested implementing classification based on molecular characterization[10], which can be useful to define targeted therapy and monitor cancer development and treatment. Moreover, there are currently no targeted therapies: in fact, there is no molecular target for treatment, detection, or monitoring. Finally, 75% of patients have early-stage, low-grade endometrial endometrioid carcinoma (EEC), which can be treated by surgery; however, about 15% of patients develop recurrence, which cannot be correctly predicted at diagnosis[11].
In this paper, we investigate the classical biomarkers in blood LB and the new ones in other biofluids for EC [Table 1] by reviewing the data in journal databases.
Liquid biopsy biospecimen and molecules in endometrial cancer
| Biospecimen | Biomolecule | Biomarker | Suggested for | Not assayed in EC | Sample analyzed | Ref. |
|---|---|---|---|---|---|---|
| Bloodn | CTCs | Protein
mRNA | Diagnosis | 10
62 24 92 24 311 | [81]
[24] [28] [29] [30] [31] | |
| DTCs | DNA | Diagnosis | 34
401 13 24 | [22]
[24] [27] [28] | ||
| Single CTC | x | [30] | ||||
| Ct/cfDNA | Mutation | Diagnosis | 44
109 48 5 | [37]
[50] [51] [52] | ||
| Integrity/
amount | Biomarkers | 60
53 | [53]
[55] | |||
| CNV | 53 | [54] | ||||
| mtDNA | Presence | 59 | [56] | |||
| miRNA | Expression | Diagnosis/prognosis | 3580
44 32 77 42 267 93 3568 | [24]
[63] [64] [66] [67] [72] [73] [75] | ||
| Methylation | Biomarkers | 45 | [76] | |||
| mRNA | Expression (hTER) | Biomarkers | 56 | [79] | ||
| Protein | Expression (leptin) | Biomarkers | 10 | [77] | ||
| HE4 | Biomarkers | 5 | [78] | |||
| circRNA | x | [101] | ||||
| lncRNA | x | |||||
| snRNA | x | |||||
| snoRNA | x | |||||
| piRNA | x | |||||
| Extracellular vesicle | DNA | x | ||||
| miRNA | x | |||||
| mRNA | x | |||||
| circRNA | Biomarkers | 3 | [84] | |||
| Protein | Biomarkers | 10 | [81] | |||
| Platelet | x | [85] | ||||
| miRNA | x | |||||
| mRNA | x | |||||
| circRNA | x | |||||
| Protein | x | |||||
| lncRNA | x | |||||
| Urine | Ct/cfDNA | x | ||||
| miRNA | Biomarkers | 20 | [24,88] | |||
| mRNA | x | |||||
| Protein | x | |||||
| circRNA | x | |||||
| Extracellular vesicle | DNA | x | ||||
| miRNA | x | |||||
| mRNA | x | |||||
| circRNA | x | |||||
| Protein | x | |||||
| Uterine/peritoneal lavage | Cells (CTC) | Biomarkers/prognosis | 2 | [89] | ||
| Ct/cfDNA (uterine) | Biomarkers/prognosis | 382
5 1 107 | [24]
[96] [97] [98] | |||
| miRNA | x | |||||
| mRNA | x | |||||
| circRNA | x | |||||
| Protein | IL11 (uterine) | Biomarkers/prognosis | 16 | [95] | ||
| Extracellular Vesicle | DNA | x | ||||
| miRNA (Exosome) | 25 | [99] | ||||
| mRNA | Biomarkers | 25 | [94] | |||
| circRNA | x | |||||
| Protein | x | |||||
| Cytology (peritoneal) | Diagnosis/prognosis/screening | 46
5 22 198 30 25 | [89]
[90] [91] [92] [93] [94] | |||
Materials and methods
The evaluation of papers was performed in PubMed using the following key words: “miRNA”, “circulating miRNA”, “cell-free DNA/ctDNA”, “lncRNA”, “vesicles/exosomes”, “messenger RNA (mRNA)”, “circular/circRNA”, and “CTC/circulating Tumor cells” “LB” (blood, urine, saliva, and peritoneal lavage/fluid); AND “Endometrial cancer/gynecological malignancies”. The search retrieved 130 articles from 2015 to 2020, of which about 100 were included in this review.
Blood-liquid biopsy
When Liquid biopsy is cited in relation to tumor, the main evaluated biomarkers are CTCs and ctDNA in blood. There is no consensus when comparing ctDNA, CTCs, and primary tumor[12]; the results change depending on the tumor analyzed and the procedure adopted for biomarkers isolation and characterization. For example, these results include 92% for KRAS in colon cancer comparing ctDNA and primary tissue[13], “good correlation” between CTs and ctDNA in colon[14], and “higher accuracy of ctDNA as prognostic biomarker” in comparison to CTC in overall evaluation in cancer[15].
Concerning EC, no studies have been reported comparing CTCs, but an interesting study compares the somatic mutation profile evaluated on ctDNA extracted from plasma to that derived from peritoneal lavage on 50 patients by NGS, which shows a concordance on KRAS of 42% between primary tissue and peritoneal lavage ctDNA and 18% for PIK3CA; for plasma ctDNA, this concordance decreases to 7% and 4%, respectively[16].
Circulating tumor cells
CTCs were discovered as early as 1869 by Thomas Asworth, who found tumor cells in the blood of a person who died from metastatic cancer. CTCs were mainly investigated in prostate, breast, lung, and thyroid carcinomas. Some studies are based on the identification of cells, while other on the characterization of CTCs by the identification of specific biomarkers related to primary cancer. There are several methods for identification of CTCs:
(1) Immunoaffinity: This method uses specific antigens present on the surfaces of CTCs but not expressed on other cells. In the negative enrichment, the CD45 antigen is generally targeted to capture normal cells. In the positive enrichment, these antigens are used for identification and separation of CTCs from other blood cells. Usually, these antigens are represented by EpCAM, but they may result in false-positives; thus, it is also important to consider epithelial-to-mesenchymal transition (EMT) and stem cell markers. To overcome this challenge and to include more CTC subpopulations during separation, multiple surface markers such as EGFR, human epidermal growth factor receptor 2 (HER2), mucin 1 (MUC1), and CXCR4-SDF[17] have recently been developed as specific biomarkers related to CTC-plasticity phenotype, stemness, and epithelial-to-mesenchymal transition (EMT) features that may provide an advantage in the promotion of metastasis for CTC dissemination and homing (by the CellSearch system and CTC-chip and ISET)[18] or to set immunotherapy[19]. Even if they can be related to specific surface proteins characterizing the primary tumors, there is a chance that they may be lost during tumor development in some tumor clones. Two of the main disadvantages and challenges of immunoaffinity-based CTC isolation methods can be addressed to the heterogeneity of CTCs, which can cause a loss of CTC subpopulations.
(2) Immunomagnetic positive enrichment: Immunomagnetic strategies are based on the previous strategy and the antigens for CTC identification are bound to magnetic beads (not to a surface as for immunoaffinity), even if this can cause a reduction in the CTC capture efficiency and result in CTC loss. Consequently, this technology has recently been improved with microfluidic and nanoparticles (e.g., graphene oxide, silica nanoparticle, and gold nanoparticle-thiol exchange reaction).
(3) Size-based technique: It is a physical method based on cell-sized: usually, CTCs have a higher size (9-19 μm) in comparison to other blood cells (about 8 μm). Sized-based techniques use membrane microfilters (typically polycarbonate films containing controlled nano- to micron-sized pores) and microfluidics under controlled pressure to prevent mechanical damage to cells during filtration. This method avoids the inaccuracy of heterogeneous antigen expression observed in CTCs. The main advantage of this technique is that it is label-free (e.g., ISET, ScreenCellCyto, and Parasortix).
(4) Membrane capacitance: Dielectrophoretic field-flow fractionation (DEP-FFF) employs separation by size and polarizability using membrane capacitance, which can process 30 million cells within 30 min with high recovery rates. However, it requires very specific parameters such as cell type and electric field frequency (e.g., DEPArray)[20].
(5) Density based: This procedure is based on centrifugation, which uses the specific density of RBCs, leukocytes, and cancer cells on specific buffer. This method is one of the first reported.
(6) Reactive ion etching: This procedure is based on photolithography to make specific patterns on the glass surface that favor CTC attachment to normal cell attachment on the basis of adhesion preferences[20].
(7) Acoustic-wave fields inside microchannels are used to capture CTCs based on the principle that all cells experience different acoustic radiation forces that result in varying movement trajectories, ultimately separating the cells. In most cases, this procedure is associated with others, including size, density, and compressibility.
(8) Combined method: It is the combination of more than one of the above-listed procedures (e.g., CTC-iChip1 and CTC-iChip2)[21,22].
All the aforementioned technologies have been designed for cell capture ex vivo. Nowadays, a new technology, known as GILUPICellCollector®, applies an anti-EpCAM wire directly into the peripheral arm vein and captures CTCs with remarkable efficiency, processing approximately 1.5 L of blood in 30 min[20].
A new definition was recently introduced related to classical CTCs. It is not known if all CTCs are able to induce relapse or metastasis. Consequently, the term CTCs refers to tumor cells that can be found in circulation. Those CTCs able to induce metastasis and disseminate are defined disseminated tumor cells (DTCs) based on the presence of specific antigens on their surface, identified by immunocytochemistry (ICC) or via their corresponding mRNA by reverse transcription quantitative polymerase chain reaction (RT-qPCR)[21,23]. Generally, CTCs, after enrichment, are evaluated to identify specific cancer-related profile biomarkers by fluorescence in situ hybridization for genome amplification detection, ICC for protein markers identification, and RT-PCR/RT-qPCR/NGS for quantifying specific RNA and DNA sequence analysis[20]. It is necessary to underline that most of the procedures for CTC isolation, characterization, and count were designed for cancers other than EC, therefore they may not be appropriate for recognizing specific antigens related to EC-CTC.
Only some of the above-mentioned technologies for CTC isolation are currently approved for in vitro diagnostics and are mainly not specific for EC. These technologies may not be suitable to properly identify EC-CTCs and could give different results. Moreover, these techniques may be inappropriate to identify different biomarker profiles based on DNA mutations[24], DNA methylation[25], mRNA expression[26-28], or protein[28].
Count of CTCs
There are few articles related to CTC count in EC. One is based on CTC enumeration by MetaCell® technology, which has been detected in 75% of patients with EC[29]. Another one is based on the identification of circulating progenitor cell number (characterized by CD34, VEGFR2, and KDR expression) in the PB of women with early EC. Those numbers result significantly augmented compared with the ones coming from healthy control women[24]. Another study evidenced that the detection of more than two CTCs is useful for a preoperative diagnosis of grade 3 EC: patients with poorly differentiated endometrioid EC have higher CTC number. As suggested by Bogani et al.[30], the discrepant results are mainly due to different biomarkers used for CTC characterization. Regarding DTCs, it has been reported that they are present in 16% of bone marrow aspirates of women affected by EC[31].
Biomarkers of CTC
Some studies focused on the identification of the expression of single specific biomarkers such as Mig7, CK19[27], and thyroid transcription factor-1, which were found to be correlated with TNM staging, vascular infiltration, lymphatic metastasis, progression-free survival, and the decrease of median survival time; consequently, they are proposed as good markers for endometrial carcinoma and metastasis[26]. Other studies have proposed mRNA multi-markers based on the isolation of CTCs or by CellSearch followed by RT-qPCR analysis. Bao et al.[28] proposed the following pan: CK20, CEA, AGR2, MGB2, DLL4, EphA2, Her3, and PDGFRα. Obermayr et al.[32] proposed another pan focused on: CCNE2, DKFZp762E1312, EMP2, MAL2, PPIC, and SLC6A8.
Another study explored the presence of specific mutations EC-related genes such as CTNNB1, STS, GDF15, RELA, RUNX1, BRAF, and PIK3CA, which may be suggested as potential therapeutic targets[24].
A consensus panel profile based on DNA or mRNA CTC analysis has not been defined, due to the low number of studies performed, the different technologies used for CTC isolation and biomarker analysis, and the different biomarkers analyzed and procedures used for identification. Moreover, another challenge is represented by the need of specific databases and algorithms able to integrate several sources of information derived from analyzing multiparametric data as well as multiple biospecimens (e.g., CTC and ctDNA)[33].
Count and biomarkers of CTCs
Several studies have analyzed both the count of CTCs and their molecular characterization, in which the antigen for isolation was used as a marker as well as for enumeration, e.g., EpCAM and stathmin [as confirmed by immunohistochemistry (IHC)], showing an increase in CTC number and stathmin IHC in non-endometrioid versus endometrioid histology, tumor size ≥ 5 cm vs. < 5 cm, higher-stage disease, and worse survival[28].
An investigation performed on patients affected by EC with and without recurrence, ranging from Grade 3 Stage IB to Stage IV carcinomas and recurrences, by EpCAM-based immunoisolation using the CELLectionTM Epithelial Enrich kit (Invitrogen, Dynal) followed by RT-qPCR analysis, associated the presence of CTC with high-risk EC, evidencing the CTC-plasticity phenotype with stemness and EMT gene-expression profile[22].
CTC in vitro for drug response model
The ability to isolate live CTCs by specific device (e.g., MetaCell®), as reported by Kolostova, might be useful to test in vitro specific drug treatments[34].
Single CTC analysis
Although limited in sample size and number of studies, due to the highly technical and expensive procedures, for characterizing the profile of CTCs, it may be useful to perform genetic and expression profiles of single CTC, which have demonstrated superior diagnostic accuracy in defining lineage identity in other tumors such as multiple myeloma and prostate cancer[35]. The main procedure adopted is the evaluation of genetic profile by NGS after whole genome amplification[20].
ctDNA
Biology of ctDNA
The presence of ctDNA was first described more than 30 years ago, even if intensive research began only in the 2010s. Active and passive mechanisms were proposed for the origin of cell-free DNA (cfDNA). Through passive mechanism, cfDNA is released into the blood via apoptosis and necrosis performed by macrophages and phagosomes from hematopoietic cells. Through active mechanism, tumor cells secrete cytoplasmic fragmented DNA to communicate with distant tissue through exosomes.
The fragment size of cfDNA is 170-200 bp, reflecting the structure of nucleosomes, sustained by nuclease digestion of genomic DNA (gDNA) during apoptosis. The fragment size profile differs between cfDNA and ctDNA from tumor cells. Shorter (< 100 bp) or longer (> 10,000 bp) fragments are frequently observed in ctDNA from cancer patients as the majority of the cfDNA from normal cells is released after apoptosis, while ctDNA can also be released by necrosis. Necrosis-based fragmentation appears to create longer or shorter DNA fragments in comparison to physiological apoptotic origin in healthy subjects. Short ctDNA fragments are more frequently observed in patients with metastatic disease than in those with early-stage cancer.
After surgical resection, the half-life of ctDNA was estimated to be 114 min. The cfDNA amount was estimated to be three-fold higher in cancer patients compared with that in healthy individuals[36]. However, pregnancy and some disorders, such as infectious and autoimmune diseases, stroke, infarction, and trauma, induce an increase in cfDNA[37]. The ctDNA amount shows wide variation among cancers, differs among stages, and may correlated with tumor burden[36]. The evaluation cfDNA in term of amount and integrity as well as studying specific mutations is cheaper and easier than CTC evaluation.
ctDNA has already been implemented in routine clinical practice after European Medicines Agency approval of the EGFR mutation test (Therascreen EGFR Plasma, Qiagen) in plasma of patients with non-small cell lung cancer[24].
Nevertheless, no consensus or standardized analytical procedures have been defined to harmonize cfDNA procedures (although there is agreement concerning the isolation of cfDNA and ctDNA from plasma, being serum preferred) or the analytical phase (methodologies, reference genes for integrity, and total amount evaluation).
Levels of cfDNA in serum are higher than in plasma due to contamination of gDNA from leucocytes during the clotting process; therefore, plasma is preferable. For the same reason, cfDNA needs to be isolated from standard EDTA-blood tubes within 2-4 h. Alternatively, tubes containing stabilizers that prevent cell lysis should be used for blood collection[38]. However, two centrifugations should be performed, the first within 2-4 h for separating plasma from whole blood at 1000-2000 g and 4 °C for 10-15 min and the second performed on plasma to separate plasma form platelets or cellular debris for the same time and temperature but at about 15,000 g[38]. ctDNA assays can identify tumor-specific genetic alterations (e.g., somatic point mutations, loss of heterozygosity, gene fusions, gene copy number variations, and DNA methylation changes)[3,36,39,40], allow the monitoring of cancer evolution and the setting of specific personalized targeted therapy[41], and provide suggestions for immunotherapy[42]; they can allow detect minimal residual disease, monitor mutations that are related to tumor burden, and define the appearance of resistance to targeted therapy[43].
Methodologies for detection of ctDNA
The methodologies depend on the purpose of ctDNA[44-48]:
(1) Quantitative PCR (qPCR): This procedure is used for the evaluation of total cfDNA amount and/or cfDNA integrity. To be sure to identify properly ctDNA coming from EC (and not from others tumors or pathologies), it is necessary to refer to specific mutations present in the tumor. Instead, for the evaluation of total cfDNA amount and integrity, it is necessary to use a specific gene that is not amplified or differentially fragmented in other tumors or pathologies.
(2) ddPCR: This procedure is mainly related to the evaluation of the presence of specific tumor-related mutations. The concept of this procedure is based on Poisson distribution and allows the absolute quantification of each allele or mutation considering the number of wells containing the target genes in comparison to wild type (wt) taking into consideration that a single molecule is present in each well. Standard PCR or qPCR instead evaluate the median distribution of a mixed mutated and wt solution, which may decrease the accuracy and sensitivity for the detection of rare mutations. Several instruments allow ddPCR, which can be distinguished depending on the size of generated droplets.
(3) NGS: This procedure is mainly related to the evaluation of the presence of specific tumor-related mutations, although it can also be used to evaluated copy number variation (CNV) and quantification of total amount. Deep massive paralleling allows the evaluation of multiple mutations (several mutations in the same gene and several genes) and more than one sample in the same run[49].
Mutations of ctDNA
Several studies have evidenced the presence of ctDNA in gynecological malignancies and the detection of specific mutations related to EC.
One of the first studies was performed by Dobrzycka et al.[50], using PCR-restriction fragment length polymorphism in a cohort of 109 patients with EC to analyze TP53 and KRAS mutations, confirming the data reported for primary tumors that there is a higher percentage of TP53 mutation in serous carcinomas as well as a high frequency of KRAS mutations in grade 2 endometrioid tumors. Pereira et al.[37] observed the presence of mutations in TP53 PTEN, PIK3CA, MET, KRAS, FBXW7 and BRAF in ctDNA, and these detections were useful to predict the tumor recurrence, with an average of seven months, before the radiologic evidence.
Moreover, Bolivar et al.[51] evidenced the presence of mutations on CTNNB1, KRAS, PTEN, or PIK3CA genes, independently of total cfDNA amount, in association with advanced stage, deep myometrial invasion, lymphatic/vascular invasion, and primary tumor size.
As discussed below, even for ctDNA of EC, there is evidence of discrepancy between the mutations found in plasma compared to those evaluated in primary tissue sample with an agreement of only 33%[52].
Integrity index of ctDNA
Only one study has evaluated the integrity index performed on ctDNA extracted from serum and based on evaluation of Alu repeats. The authors registered an increase in ctDNA integrity and amount in high grade compared to G1 ECs, suggesting a role of cfDNA as potential prognostic biomarker in EC[53].
Copy number variation in ctDNA
The same findings were obtained for the evaluation of CNV in ECs by NGS by Nakabayashi et al.[54] on three patients on the following loci: 1p36-p31 and 1q12-q44 or 8q24.
Total amount of cell free DNA
Regarding the total cfDNA evaluation, the two reported articles are based on the evaluation of Alu repeats: the previous citation[53] suggests a relationship between integrity and EC grade, while another[55] reports no significant difference in cell-free DNA among stage or histological grade of EC, as well as no significant change in cell-free DNA before and after operation.
Mitochondrial ctDNA
Recent studies have demonstrated a potential link between circulating mitochondrial cell-free DNA (cfmtDNA) content and cancer. In particular, the evaluation of aberrant changes and altered content of cfmtDNA represents an important approach for early cancer diagnosis with some unique advantages over circulating nuclear cell free DNA (cfnDNA), such as the much shorter and more simply organized mitochondrial genome and the higher number of mtDNA copy. These characteristics make screening much easier and more cost-effective, with respect to the nuclear genome one, in body fluid samples such as plasma and serum. mtDNA does not have protective histones and sophisticated DNA repair mechanisms, which underscore its susceptibility to oxidative stress and other genotoxic insults. The abnormal alteration of mtDNA copy number is well documented for numerous malignancies. Recently, Cicchillitti et al.[56] evidenced an association among cfmtDNA (by qPCR), EC grading, and hypertension and inflammation markers, suggesting its role as a predictive biomarker[57-62].
miRNA
miRNA expression
miRNAs are involved in the pathogenesis of various human cancers, such as lung, prostate, colorectal, and leukemia, as either oncogenes or onco-suppressors. The main emerging evidence of miRNAs is related to their use as biomarkers due to their higher stability in comparison to longer RNAs and their relatively high concentration in body fluids, such as serum, plasma, saliva, and urine, initiating a new era of disease research.
Circulatory miRNAs are reported mainly from monocytes, plasma, and exosomes, and they are resistant to degradation by RNase enzyme, thus stable in the blood and urine. Consequently, the potential roles of miRNAs in clinical practice are related to early diagnosis and classification of tumors, in the identification of poorly differentiated malignancies, and in the use as biomarkers potential use in the prediction of survival and response to treatment.
Recent studies have expanded our knowledge of the roles of miRNA in the pathology of gynecologic malignancies: in ovarian cancer, miRNAs participate in the development of drug resistance, while, in EC, they play essential roles in oncogenic processes, including cell proliferation, migration, and metastasis. The most critical aspect, more than in CTCs and ctDNA, is the lack of standard procedures for the analytical and pre-analytical phases (e.g., the absence of housekeeping genes for relative quantification) that leads to the identification of a broad range of miRNAs but none of them with strong confirmation for clinical use.
Tan et al.[63] showed a higher expression of the serum level of miR-155 in differentiated EEC in comparison to healthy controls and associated it with cancer stage, lymph node involvement, and metastasis. Zhai obtained the same results for miR-194[64]. Torres found that the expression of miR-99a, miR-100, and miR-199b was upregulated in plasma of EEC patients, and a combination of miR-99a and miR-199b was more accurate in distinguishing EEC disease when compared with single miRNAs[65]. Another genome-wide serum miRNA expression profile identified a combination of four serum miRNAs (miR-222, miR-223, miR-186, and miR-204) as a fingerprint for EEC detection[66].
Other investigations of miRNA profiling in EEC have tried to find associations between circulating miRNAs and clinic-pathological characteristics such as FIGO stage, grade, relapse, and nodal metastases. The miRNAs expression is mostly linked to that in corresponding tumor tissue[24,67-72]. In this regard, Wang et al.[73] found that miR-15b, -27a, -223, miR-3145, and miR-4638, obtained by genome-wide miRNA expression profiles, are differentially expressed in the EEC plasma between the two cohorts[73,74].
Nevertheless, only one meta-analysis was performed on the discovered miRNAs related to EC. However, inconsistencies or discrepancies about diagnostic accuracy of circulating miR-21 remain. EC patients showed higher miR-21 expression compared with benign lesion patients[75].
miRNA methylation
Mainly focusing on their role as biomarkers, several miRNAs have been explored for their methylation profile in tissue and their correspondent expression in serum, but it has only been confirmed for miRNA-203[76].
Protein circulating than CA125
Only two studies investigated proteins related to EC. Tessitore et al.[77] reported an increase of circulating leptin in gynecological malignancies and breast cancer related to the increase of corresponding mRNA in primary tumor tissue. This increase was related to cachexia and hormonal markers such as estrogen receptor (ER) and progesterone receptor and, only in post-menopause, to an increase in circulating estradiol.
Qu et al.[78] evidenced the increase of serum epididymis protein 4 (HE4) level and suggested its use as a biomarker for the management of ovarian and endometrial cancer patients.
mRNA
Recently, specific mRNAs have been investigated in LB of EC patients. Few studies have been conducted, and a single article reports that hTERT mRNA was detected only in cancer patients in comparison to healthy subjects and that levels increased with tumor stage[79].
Extracellular vesicles
Extracellular micro- and nano-membrane vesicles produced by different cells progressively attract the attention of the scientific community. They function as mediators of intercellular communication transporters of genetic material and signaling molecules between cells. In the context of keeping homeostasis, extracellular vesicles contribute to the regulation of various systemic and local processes. Exosomes, microvesicles, also referred to as microparticles or ectosomes, and large oncosomes were defined as actively released vesicles.
Because extracellular vesicles (EVs) contents reflect the contents of the cell of origin, multiple studies on EVs from body fluids, in the context of cancer diagnosis, prediction, and prognosis, have been performed. EV-based LB provided an overview of the main EV constituents as potential biomarkers: surface proteins, intravesicular soluble proteins, lipids, DNA, and RNAs, including mRNA (intact and fragmented), miRNA, piwiRNA (piRNA), transfer RNA (tRNA), fragments of ribosomal RNA (rRNA), long noncoding RNA (lncRNA), and circular RNAs (circRNAs)[80].
EVs have been identified as the main mediators of cell-to-cell communication between tumor and stromal cells in local and distant microenvironments participating in the formation of the premetastatic niche prior to CTC colonization[81]. EVs can be involved in the response to immunotherapy[82], and, moreover, they may be used as drug delivery systems[83]. They can be isolated from several biospecimens such as blood, urine, CSF, lymphatics, tears, saliva, nasal secretions, ascites, and semen.
Few studies have dealt with the investigation of EV levels in EC, and these studies were performed on circulating EVs. In particular, Xu et al.[84] explored the role of circRNAs in EVs isolated from the serum of affected patients. They found 275 circRNAs to be differentially expressed, among which 209 were upregulated and 66 downregulated. All circRNAs have been identified thanks to wide-expression analysis, and two of them (hsa_circ_0109046 and hsa_circ_0002577) were confirmed by RT-qPCR.
An interesting article evidenced in advanced EC, via CellSearch® technology, the presence of CTCs in high-risk EC patients. Those CTCs were characterized by an EMT-expression profile correlated with the increase of EVs that have been found containing extracellular-matrix-like collagens (COL18A1), proteoglycans (VCAN, AGRN, and HSPG2), glycoproteins (TNC), constituents of the cytoskeleton (ACTG1, TUBA1B, and TUBB), integrins (ITGA3), and laminins (LAMA5). The above-cited proteins are all associated with tumor cell adhesion to the endothelium and to the promotion of their adhesion and colonization at distant sites. In particular, the authors found, by means of targeted proteomics, that the adhesion protein LGALS3BP was significantly enriched in circulating EVs from an EC patient cohort with a high risk of recurrence[81].
Platelet
Platelets contain many different RNA species including miRNAs, circRNAs, and mRNAs that are altered in cancer. No studies regarding platelets have been performed on EC[85].
Urine
miRNA
The improvement of high throughput technologies such as NGS and qPCR allowed the evaluation of specific biomarkers, such as cell-free miRNAs, in urine[86] and methylation profile of cell free tumor[87].
A study reported a specific downregulation of miR-106b, as well as in serum and plasma samples, in comparison with healthy donors[24].
Recently, Ye reported the identification of several miRNA involved in Grade 3 EC (miR-9, miR-92a, miR-99a, miR-100, miR-199b, miR-1228, miR-9, miR-1228, miR-9, miR-92a, miR-21, miR-222, miR-223, miR-186 and miR-204, miR-203, miR-21, miR-887-5p, miR-106b, and miR-200c-3p)[88].
Uterine/peritoneal lavage cytology
Surgical staging of gynecologic neoplasms, mainly in ovarian cancer, include the collection of peritoneal washings coming from the abdomen and pelvis. The aim of taking peritoneal washings is to identify occult disease. Peritoneal cytology is supposed to add information on the spread of microscopic peritoneal disease. However, the peritoneal washing cytology examination may give false positivity in benign diseases and false negativity in the early stages.
Less is known about the prognostic impact of peritoneal cytology in EC and published data show inconsistent results, mainly in relation to the prognostic importance of positive cytology to predict relapse or metastasis[89]. Recently, the use of cytology PAP-test, usually performed for cervical cancer evaluation, has been proposed to analyzed endometrial cancer, in particular the liquid cytology approach (LC)[90]. This procedure is worth mentioning in this overview, even if this procedure is not a true LB but a procedure in between biopsy and LB, mainly because the collection procedure is minimally invasive and allows collecting tumor cells derived from gynecological districts. After uterine lavage with saline solution, the collection of cancer cells can allow identifying genetic variations related to endometrial cancer by the washing of the uterine cavity. The data suggest that LC is a feasible and reproducible adjuvant method for screening endometrial lesions and in combination with classical biopsy can improve the diagnostic accuracy of endometrial lesions[90-94].
To identify other biomarkers for implementing cytology, recent studies demonstrated the role of IL-11 protein in uterine fluids related with the amount in endometrial tumor epithelial cells in women with grade 1 EC[95]. In a case report, in cells deriving from uterine lavage, mutations on PTEN, TP53, PIK3CA, PIK3R1, KRAS, CTNNB1, FGFR2, RNF43, PPP2R1A, POLE, APC, and FBXW7 were observed; these are genes that are some of the most frequently mutated in EC[24,96-98]. In addition, exosome contents, mainly referring to miRNAs, showed that 114 miRNAs (by mRNA array) were significantly dysregulated in EC patients. Eight miRNAs (miRNA-383-5p, miRNA-10b-5p, miRNA-34c-3p, miRNA-449b-5p, miRNA-34c-5p, miRNA-200b-3p, miRNA-2110, and miRNA-34b-3p) demonstrated classification performance according to the area under the receiver operating characteristic curve[99].
lncRNA and circRNA and other small nuclear/nucleolar RNA in LB
There are currently no studies available relating other circulating RNAs, namely lncRNA, piRNA, small nuclear (snRNA), and small nucleolar (snoRNA), to EC. The last three classes are also known as snc/snRNAs due to the fact that they are located in the cell nucleus and are fundamental in RNA-RNA remodeling, spliceosome assembly, and translation processes (i.e., post-transcriptional modification of rRNA). The class of snoRNAs named “U(n)” (i.e., U1 and U2-U12), due to their high Uridyl content, is involved in the spliceosome complex. These particular RNAs are 60-300 nt long and are transcribed from intronic sequences of coding and noncoding genes. Some of them are used to normalize the relative quantification of miRNA in tissues and fluids. Although they do not properly represent “housekeeping” due to their change and different pre-analytical phase, no specific studies have been conducted on these biomolecules to evaluate their possible function as biomarkers[88].
Concerning lncRNAs, they are involved in chromatin remodeling, gene expression and transcription, and protein-protein interaction, and their role in cancer is known with respect to snRNAs. Since this is a new field, for EC, pilot studies concerning circulating lncRNA are only relative to colon cancer and their prospective use[100].
Noteworthy, a peculiar class of lncRNA is represented by circRNAs characterized by a covalent linkage, which gives them a specific circular form and makes them biologically stable and resistant to RNases. They were recently investigated in several cancer tissues including EC. A recent paper highlights their differential display in a pilot study aiming to analyze three samples of EC in comparison to corresponding adjacent non-cancerous tissue and focused on has-circ_0039569, which has been significantly correlated with tumor differentiation[88].
Recently, circRNAs - hsa_circ_0109046 and hsa_circ_0002577 - were suggested as potentially investigable biomarkers in LB of EC[101].
Role of LB in the monitoring and treatment of endometrial cancer
Patients with EC progression after first-line chemotherapy have a poor prognosis. Until now, no targeted therapies are in use for ECC treatment, although several phosphoinositide 3-kinase inhibitors are under investigation and in clinical trials. As for lung cancer, the evaluation of specific circulating mutations may give information concerning the success of the therapy and related to the development of specific molecular resistance or new clones[102]. Several studies have found that the genotyping results derived from tissue biopsy analysis differ from those derived from LB. The concordance rates for metastatic cancer patients and for patients with primary tumors were 83.3% and 78.3%, respectively. These discrepancies may be related to intra-tumor heterogeneity, indicating that the assessment on tissue can cause misinterpretations, while LB may reflect mutations and changes occurring in tumor that cannot be revealed in the primary biopsy. Thereby, LB may offer new prospective for monitoring tumor development, the efficacy of therapy, and the arise of treatment resistance[36].
Conclusion
In 2013, the Cancer Genome Atlas Research Network (TCGA) suggested implementing a classification with molecular characterization[10], which can be useful to define targeted therapy and monitor cancer development and treatment; nevertheless, today, there are no targeted therapies: in fact, there is no molecular target for treatment, detection, or monitoring[11]. LB represents a novel tool, due to the minimally invasive- or non-invasive procedure for biomarker collection, overcoming the limit of classical tissue biopsy and allowing the monitoring of tumor burden, the efficacy of therapy, the arise of resistance, and the development of cancer change, relapse, and metastasis.
As discussed in this review, the investigation of biomarkers in LB of EC evidenced that only experimental approaches have been proposed, and none of them have reached clinical application. In particular, most of the results concerning LB in EC are referred to circulating cell free DNA/ctDNA and CTCs in blood. This fact may be related to the statement that most ECs have a good prognosis and are diagnosed at early stage, and thus little effort has been made in this direction despite the improvement of new technologies (e.g., NGS and ddPCR) and new isolation systems for emerging biospecimen for LB in cancer detection (e.g., CTC and EV) The overview of LB biomarkers in EC shows that blood is the most investigated biospecimen and the main biomolecules are miRNA and ctDNA. Nevertheless, for those biomarkers evaluated in EC, some discrepancies were observed among studies, mainly related to the lack of standard procedures for pre-analytical and analytical phases. This aspect plays a critical role in EC, principally due to the limited number of studies or patients recruited within each study performed on this topic. Nevertheless, to underline the efforts performed in this direction, in this review, we present, albethey isolated, studies that for the first time performed analyses of specific biomarkers in LB of EC (e.g., circulating mRNA in blood) and we provocatively suggest to extend them to biomarkers (lncRNA, methylation of circulating miRNA, and single CTC analysis) not yet analyzed in EC but investigated in other cancers. The purpose is to suggest that LB of EC represents a fascinating tool and field of study to improve the knowledge of this type of tumor and to improve the life-quality of patients.
Declarations
Authors’ contributionsMade substantial contributions to conception and design of the study: Malentacchi F, Pillozzi S
Data research: Malentacchi F, Sgromo C
Revision of the manuscript: Antonuzzo L
Availability of data and materialsNot applicable.
Financial support and sponsorshipNone.
Conflicts of interestAll authors declared that there are no conflicts of interest.
Ethical approval and consent to participateNot applicable.
Consent for publicationNot applicable.
Copyright© The Author(s) 2020.
REFERENCES
1. Aldana Blanco B, Langenbeck’s CLW. Liquid biopsy for the detection and management of surgically resectable tumors. Arch Surg 2019;404:517-25.
2. Hyun KA, Gwak H, Lee J, Kwak B, Jung H. Salivary exosome and cell-free DNA for cancer detection. Micromachines 2018;9:340.
3. Rossi G, Ignatiadis M. Promises and pitfalls of using liquid biopsy for precision. Med Cancer Res 2019;9:2798-804.
4. Geeurickx E, Hendrix A. Targets, pitfalls and reference materials for liquid biopsy tests in cancer diagnostics. Mol Aspects Med 2020;72:100828.
6. Morice P, Leary A, Creutzberg C, Abu-Rustum N, Darai E. Endometrial cancer. Lancet 2016;387:1094-108.
7. McGuire S. World cancer report 2014. Geneva, Switzerland: world Health organization, international Agency for research on cancer, WHO press. Adv Nutr 2015;7:418-9.
9. Levine DA. The Cancer Genome Atlas Research Network Integrated genomic characterization of endometrial carcinoma. Nature 2013;497:67-73.
10. Colombo N, Creutzberg C, Amant F, Bosse T, González-Martín A, et al; ESMO-ESGO-ESTRO Endometrial Consensus Conference Working Group. ESMO-ESGO-ESTRO Consensus Conference on Endometrial Cancer: diagnosis, treatment and follow-up. Ann Oncol 2016;27:16-41.
11. Ruz-Caracuel I, Ramón-Patino JL, López-Janeiro Á, Yébenes L, Berjón A, et al. Pattern as a prognostic marker in low-grade, early-stage endometrioid endometrial carcinoma. Cancers 2019;11:e1845.
12. Marius Ilié M, Hofman P. Pros: Can tissue biopsy be replaced by liquid biopsy? Transl Lung Cancer Res 2016;5:420-3.
13. García-Foncillas J, Tabernero J, Elena Élez E, Enrique Aranda E, Benavides M, et al. Prospective multicenter real-world RAS mutation comparison between OncoBEAM-based liquid biopsy and tissue analysis in metastatic colorectal cancer. Br J Cancer 2018;119:1464-70.
14. Bidard FC, Kiavue N, Ychou M, Cabel L, Stern MH, et al. Circulating tumor cells and circulating tumor DNA detection in potentially resectable metastatic colorectal cancer: a prospective ancillary study to the unicancer. Prodige-14 Trial. Cells 2019;8:516.
15. Tuaeva NO, Falzone L, Porozov YB, Nosyrev AE, Trukhan VM, et al. Translational application of circulating DNA in oncology: review of the last decades achievements. Cells 2019;8:1251.
16. Mayo-de-Las-Casas C, Velasco A, Sanchez D, Martínez-Bueno A, Garzón-Ibáñez M, et al. Detection of somatic mutations in peritoneal lavages and plasma of endometrial cancer patients: a proof-of-concept study cancer. Int J Cancer 2020;147:277-84.
17. Mego M, Cholujova D, Minarik G, Sedlackova T, Gronesova P, et al. CXCR4-SDF-1 interaction potentially mediates trafficking of circulating tumor cells in primary breast cancer. BMC Cancer 2016;16:127.
18. Liu Q, Liao Q, Zhao Y. Myeloid-derived suppressor cells (MDSC) facilitate distant metastasis of malignancies by shielding circulating tumor cells (CTC) from immune surveillance. Medical Hypotheses 2016;87:34-9.
19. Kloten V, Lampignano R, Krahn T, Schlange T. Circulating tumor cell PD-L1 expression as biomarker for therapeutic efficacy of immune checkpoint inhibition in NSCL. Cells 2019;8:e809.
20. Sharma S, Zhuang R, Long M, Pavlovic M, Kang Y, et al. Circulating tumor cell isolation, culture, and downstream molecular analysis. Biotechnol Adv 2018;36:1063-78.
21. Bankó P, Lee SY, Nagygyörgy V, Zrínyi M, Chae CH, et al. Technologies for circulating tumor cell separation from whole blood. J Hematol Oncol 2019;12:48.
22. Alonso-Alconada L, Muinelo-Romay L, Madissoo K, Diaz-Lopez A, Krakstad C, et al. Molecular profiling of circulating tumor cells links plasticity to the metastatic process in endometrial cancer. Molecular Cancer 2014;13:223.
23. Masuda T, Hayashi N, Iguchi T, Ito S, Eguchi H, et al. Clinical and biological significance of circulating tumor cells in cancer. Mol Oncol 2016;10:e408-e417.
24. Muinelo-Romay L, Casas-Arozamena C, Abal M. Liquid biopsy in endometrial cancer: new opportunities for personalized oncology. Int J Mol Sci 2018;19:2311.
25. Pixberg CF, Schulz WA, Stoecklein NH, Neves RPL. Characterization of DNA methylation in circulating tumor cells. Genes 2015;6:1053-75.
26. Zhang Y, Qu X, Qu PP. Value of circulating tumor cells positive for thyroid transcription factor-1 (TTF-1) to predict recurrence and survival rates for endometrial carcinoma. J BUON 2016;21:1491-5.
27. Kolbl AC, Wellens R, Koch J, Rack B, Hutter S, et al. Endometrial adenocarcinoma: analysis of circulating tumour cells by RT-qPCR. Anticancer Res 2016;36:3205-10.
28. Bao H, Burke PA, Huang J, Chen X, Brohawn PZ, et al. Circulating tumor cells: application as a biomarker for molecular characterization and predictor of survival in an all-comer solid tumor phase I clinical study. PLoSOne 2013;8:e58557.
29. Kiss I, Kolostova K, Matkowski R, Jędryka M, Czekański A, et al. Correlation between disease stage and the presence of viable circulating tumorcells in endometrial cancer. Anticancer Res 2018;38:2983-7.
30. Bogani G, Liu MC, Dowdy SC, Cliby WA, Kerr SE, et al. Detection of circulating tumor cells in high-risk endometrial cancer. Anticancer Res 2015;35:683-8.
31. Walter CB, Taran FA, Wallwiener M, Rothmund R, Kraemer B, et al. Prevalence and prognostic value of disseminated tumor cells in primary endometrial, cervical and vulvar cancer patients. Future Oncol 2014;10:41-8.
32. Obermayr E, Sanchez-Cabo F, Tea MKM, Singer CF, Krainer M, et al. Assessment of a six gene panel for the molecular detection of circulating tumor cells in the blood of female cancer patients. BMC Cancer 2010;10:666.
33. Zou J, Wang E. eTumorType, an algorithm of discriminating cancer types for circulating tumor cells or cell-free DNAs in blood. Gen Prot Bioinformatics 2017;15:130-40.
34. Kolostova K, Spicka J, Matkowski R, Bobek V. Isolation, primary culture, morphological and molecular characterization of circulating tumor cells in gynecological cancers. Am J Transl Res 2015;7:1203-13.
35. Lim SB, Lee WD, Vasudevan J, Lim WT, Lim CT. Liquid biopsy: one cell at a time. NPJ Precis Oncol 2019;3:23.
36. Snyder A, Morrissey MP, Hellmann MD. Use of circulating tumor DNA for cancer immunotherapy. Clin Cancer Res 2019;25:6909-15.
37. Pereira E, Camacho-Vanegas O, Anand S, Sebra R, Camacho SC, et al. Personalized circulating tumor DNA biomarkers dynamically predict treatment response and survival in gynecologic cancers. PLoS One 2015;10:e0145754.
38. Malentacchi F, Pizzamiglio S, Verderio P, Pazzagli M, Orlando C, et al. Influence of pre-analytical phase on circulating cell free DNA (ccfDNA): the SPIDIA-DNAplas external quality assessment experience. Clin Chem Lab Med 2015;53:1935-42.
39. Nakauchi C, Kagara N, Shimazu K, Shimomura A, Naoi Y, et al. Detection of TP53/PIK3CA mutations in cell-free plasma DNA from metastatic breast cancer patients using next generation sequencing. Clin Breast Cancer 2016;16:418-23.
40. Zeng C, Stroup EK, Zhang Z, Chiu BCH, Zhang W. Towards precision medicine: advances in 5-hydroxymethylcytosine cancer biomarker discovery in liquid biopsy. Cancer Commun 2019;39:12.
41. Kerachian MA, Poudineh A, Thiery JP. Cell free circulating tumor nucleic acids, a revolution in personalized cancer medicine. Crit Rev Oncol Hematol 2019;144:102827.
42. Li L, Zhang J, Jiang X, Li Q. Promising clinical application of ctDNA in evaluating immunotherapy efficacy. Am J Cancer Res 2018;8:1947-56.
43. Vidal J, Taus A, Montagut C. Dynamic treatment stratification using ctDNA. Recent Results Cancer Res 2020;215:263-73.
44. Zhu Y, Yang T, Wu Q, Yang X, Hao J, et al. Diagnostic performance of various liquid biopsy methods in detecting colorectal cancer: a meta-analysis. Cancer Med 2020;9:5699-707.
45. Koessler T, Paradiso V, Piscuoglio S, Nienhold R, Ho L, et al. Reliability of liquid biopsy analysis: an inter-laboratory comparison of circulating tumor DNA extraction and sequencing with different platforms. Lab Invest 2020; doi: 10.1038/s41374-020-0459-7.
46. Moon SM, Kim JH, Kim SK, Kim S, Kwon HJ, et al. Clinical utility of combined circulating tumor cell and circulating tumor DNA assays for diagnosis of primary lung cancer. Anticancer Res 2020;40:3435-44.
47. Kerachian MA, Azghandi M, Javadmanesh A, Ghaffarzadegan K, Mozaffari-Jovin S. Selective capture of plasma cell-free tumor DNA on magnetic beads: a sensitive and versatile tool for liquid biopsy. Cell Oncol (Dordr) 2020; doi: 10.1007/s13402-020-00536-2.
48. Pittella-Silva F, Chin YM, Chan HT, Nagayama S, Miyauchi E, et al. Plasma or serum: which is preferable for mutation detection in liquid biopsy? Clin Chem 2020;66:946-57.
49. Sato Y, Matoba R, Kato K. Recent advances in liquid biopsy in precision oncology research. Biol Pharm Bull 2019;42:337-42.
50. Dobrzycka B, Terlikowski SJ, Mazurek A, Kowalczuk O, Niklinska W, et al. Circulating free DNA, p53 antibody and mutations of KRAS gene in endometrial cancer. Int J Cancer 2010;127:612-21.
51. Bolivar AM, Luthra R, Mehrotra M, Chen W, Barkoh BA, et al. Targeted next-generation sequencing of endometrial cancer and matched circulating tumor DNA: identification of plasma-based, tumor-associated mutations in early stage patients. Mod Pathol 2019;32:405-14.
52. Iwahashi N, Sakai K, Noguchi T, Yahata T, Matsukawa H, et al. Liquid biopsy-based comprehensive gene mutation profiling for gynecological cancer using cancer personalized profiling by deep sequencing. Sci Rep 2019;9:10426.
53. Vizza E, Corrado G, De Angeli M, Carosi M, Mancini E, et al. Serum DNA integrity index as a potential molecular biomarker in endometrial cancer. J ExpClin Cancer Res 2018;37:16.
54. Nakabayashi M, Kawashima A, Yasuhara R, Hayakawa Y, Miyamoto S, et al. Massively parallel sequencing of cell-free DNA in plasma for detecting gynaecological tumour associated copy number alteration. Sci Rep 2018;8:11205.
55. Tanaka H, Tsuda H, Nishimura S, Nomura H, Kataoka F, et al. Role of circulating free alu DNA in endometrial cancer. Int J Gynecol Cancer 2012;22:82-6.
56. Cicchillitti L, Corrado G, De Angeli M, Mancini E, Baiocco E, et al. Circulating cell-free DNA content as blood based biomarker in endometrial cancer. Oncotarget 2017;8:115230-43.
57. An Q, Hu Y, Li Q, Chen X, Huang J, et al. The size of cell-free mitochondrial DNA in blood is inversely correlated with tumor burden in cancer patients. PrecisClinMed 2019;2:131-9.
58. Pasha HA, Rezk NA, Riad MA. Circulating cell free nuclear DNA, mitochondrial DNA and global DNA methylation: potential noninvasive biomarkers for breast cancer diagnosis. Cancer Invest 2019;37:432-9.
59. Meng X, Schwarzenbach H, Yang Y, Müller V, Li N, et al. Circulating mitochondrial DNA is linked to progression and prognosis of epithelial ovarian cancer. TranslOncol 2019;12:1213-20.
60. Afrifa J, Zhao T, Yu J. Circulating mitochondria DNA, a non-invasive cancer diagnostic biomarker candidate. Mitochondrion 2019;47:238-43.
61. Mair R, Mouliere F, Smith CG, Chandrananda D, Gale D, et al. Measurement of plasma cell-free mitochondrial tumor DNA improves detection of glioblastoma in patient-derived orthotopic xenograft models. Cancer Res 2019;79:220-30.
62. Weerts MJA, Timmermans EC, van de Stolpe A, Vossen RHAM, Anvar SY, et al. Tumor-specific mitochondrial DNA variants are rarely detected in cell-freeDNA. Neoplasia 2018;20:687-96.
63. Tan ZQ, Liu FX, Tang HL, Su Q. Expression and its clinical significance of hsa-miR-155 in serum of endometrial cancer. Zhonghua Fu Chan Ke Za Zhi 2010;45:772-4.
64. Zhai H, Karaayvaz M, Dong P, Sakuragi N, Ju J. Prognostic significance of miR-194 in endometrial cancer. Biomark Res 2013;1:12.
65. Torres A, Torres K, Pesci A, Ceccaroni M, Paszkowski T, et al. Deregulation of miR-100, miR-99a and miR-199b in tissues and plasma coexists with increased expression of mTOR kinase in endometrioid endometrial carcinoma. BMC Cancer 2012;12:369.
66. Torres A, Torres K, Pesci A, Ceccaroni M, Paszkowski T. Diagnostic and prognostic significance of miRNA signatures in tissues and plasma of endometrioid endometrial carcinoma patients. Int J Cancer 2013;132:1633-45.
67. Tsukamoto O, Miura K, Mishima H, Abe S, Kaneuchi M, et al. Identification of endometrioid endometrial carcinoma-associated microRNAs in tissue and plasma. GynecolOncol 2014;132:715-21.
68. Wang WT, Zhao YN, Yan JX, Weng MY, Wang Y, et al. Differentially expressed microRNAs in the serum of cervical squamous cell carcinoma patients before and after surgery. J Hematol Oncol 2014;7:6.
69. Yu J, Wang Y, Dong R, Huang X, Ding S, et al. Circulating microRNA-218 was reduced in cervical cancer and correlated with tumor invasion. J Cancer Res Clin Oncol 2012;38:671-4.
70. Zhao S, Yao D, Chen J, Ding N. Circulating miRNA-20a and miRNA-203 for screening lymph node metastasis in early stage cervical cancer. Genet Test Mol Biomarkers 2013;17:631-6.
71. Chen J, Yao D, Li Y, Chen H, He C, et al. Serum microRNA expression levels can predict lymph node metastasis in patients with early-stage cervical squamous cell carcinoma. Int J Mol Med 2013;32:557-67.
72. Zhao YN, Chen G, Hong SJ. Circulating MicroRNAs in gynecological malignancies: from detection to prediction. Exp Hematol Oncol 2014;3:14.
73. Wang L, Chen YJ, Xu K, Xu H, Shen XZ, et al. Circulating microRNAs as a fingerprint for endometrial endometrioid adenocarcinoma. PLoSOne 2014;9:e110767.
74. La Ferlita A, Battaglia R, Andronico F, Caruso S, Cianci A, et al. Non-coding RNAs in endometrial physiopathology. Int J Mol Sci 2018;19:2120.
75. Gao Y, Dai M, Liu H, He W, Lin S, et al. Diagnostic value of circulating miR-21: an update meta-analysis in various cancers and validation in endometrial cancer. Oncotarget 2016;7:68894-908.
76. Benati M, Montagnana M, Danese E, Paviati E, Giudici S, et al. Evaluation of mir-203 expression levels and DNA promoter methylation status in serum of patients with endometrial cancer. Clin Lab 2017;63:1675-81.
77. Tessitore L, Vizio B, Pesola D, Cecchini F, Mussa A, et al. Adipocyte expression and circulating levels of leptin increase in both gynaecological and breast cancer patients. Int J Oncol 2004;24:1529-35.
78. Qu W, Gao Q, Chen H, Tang Z, Zhu X, et al. HE4-test of urine and body fluids for diagnosis of gynecologic cancer. Expert Rev Mol Diagn 2017;17:239-44.
79. Liang J, Yin G, Chen M, Wu A. Detection of hTERT mRNA in peripheral blood and its implication for diagnosis of early stage postoperative endometrial cancer micrometastasis. Panminerva Med 2016;58:206-10.
80. Nazarenko I. Extracellular vesicles: recent developments in technology and perspectives for cancer liquid biopsy. Recent Results Cancer Res 2020;215:319-44.
81. Mariscal J, Fernandez-Puente P, Calamia V, Abalo A, Santacana M, et al. Proteomic characterization of epithelial-like extracellular vesicles in advanced endometrial cancer. J Proteome Res 2019;18:1043-53.
82. Fitts CA, Ji N, Li Y, Tan C. Exploiting exosomes in cancer liquid biopsies and drug delivery. Adv Healthcare Mater 2019;8:1801268.
83. Lim W, Kim HS. Exosomes as therapeutic vehicles for cancer. Tissue Eng Regen Med 2019;16:213-23.
84. Xu H, Gong Z, Shen Y, Fang Y, Zhong S. CircularRNA expression in extracellular vesicles isolated from serum of patients with endometrial cancer. Epigenomics 2018;10:187-97.
86. Zavesky L, Jandakova E, Turyna R, Duskova D, Langmeierova L, et al. Cell-free urinary microRNAs expression in small-scale experiments. Methods Mol Biol 2017;1580:99-106.
87. Larsen LK, Lind GE, Guldberg P, Dahl C. DNA-methylation-based detection of urological cancer in urine: overview of biomarkers and considerations on biomarker design, source of DNA, and detection technologies. Int J Mol Sci 2019;20:2657.
88. Ye F, Tang QL, Ma F, Cai L, Chen M, et al. Analysis of the circular RNA transcriptome in the grade 3 endometrial cancer. Cancer Manag Res 2019;11:6215-27.
89. Binesh F, Akhavan A, Behniafard N, Zabihi S, Hosseinizadeh E. Prognostic value of peritoneal washing cytology in gynecologic malignancies: a controversial issue. Asian Pac J Cancer Prev 2014;15:9405-10.
90. Lv S, Wang R, Wang Q, Han L, Tuo X, et al. A novel solution configuration on liquid-based endometrial cytology. PLoS One 2018;13:e0190851.
91. Kinde I, Bettegowda C, Wang Y, Wu J, Agrawal N, et al. Evaluation of DNA from the Papanicolaou test to detect ovarian and endometrial cancers. Sci Transl Med 2013;5:167ra4.
92. Zhang H, Wen J, Xu PL, Chen R, Yang X, et al. Role of liquid-based cytology and cell block in the diagnosis of endometrial lesions. Chin Med J (Engl) 2016;129:1459-63.
93. Norimatsu Y, Kouda H, Kobayashi TK, Shimizu K, Yanoh K, et al. Utility of liquid-based cytology in endometrial pathology: diagnosis of endometrial carcinoma. Cytopathology 2009;20:395-402.
94. Buccoliero AM, Gheri CF, Castiglione F, Garbini F, Barbetti A, et al. Liquid-based endometrial cytology: cyto-histological correlation in a population of 917 women. Cytopathology 2007;18:241-9.
95. Yap J, Salamonsen LA, Jobling T, Nicholls PK, Dimitriadis E. Interleukin 11 is upregulated in uterine lavage and endometrial cancer cells in women with endometrial carcinoma. ReprodBiolEndocrinol 2010;8:63.
96. Maritschnegg E, Wang Y, Pecha N, Horvat R, Van Nieuwenhuysen E, et al. Lavage of the uterine cavity for molecular detection of Mullerian duct carcinomas: a proof-of-concept study. J Clin Oncol 2015;33:4293-301.
97. Martignetti JA, Pandya D, Nagarsheth N, Chen Y, Camacho O, et al. Case report: molecular detection of endometrial precancer using a targeted gynecologic cancer liquid biopsy. Cold Spring HarbMol Case Stud 2018;4:a003269.
98. Nair N, Camacho-Vanegas O, Rykunov D, Dashkoff M, Camacho SC, et al. Genomic analysis of uterine lavage fluid detects early endometrial cancers and reveals a prevalent landscape of driver mutations in women without histopathologic evidence of cancer: a prospective cross-sectional study. PLoS Med 2016;13:e1002206.
99. Roman-Canal B, Moiola CP, Gatius S, Bonnin S, Ruiz-Miró M, et al. EV-associated miRNAs from peritoneal lavage are a source of biomarkers in endometrial cancer. Cancers 2019;11:839.
100. Galamb O, Barták BK, Kalmár A, Nagy ZB, Szigeti KA, et al. Diagnostic and prognostic potential of tissue and circulating long non-coding RNAs in colorectal tumors. World J Gastroenterol 2019;25:5026-48.
101. Pardini B, Sabo AA, Birolo G, Calin GA. Noncoding RNAs in extracellular fluids as cancer biomarkers: the new frontier of liquid biopsies. Cancers 2019;11:1170.
Cite This Article
Export citation file: BibTeX | RIS
OAE Style
Malentacchi F, Sgromo C, Antonuzzo L, Pillozzi S. Liquid biopsy in endometrial cancer. J Cancer Metastasis Treat 2020;6:34. http://dx.doi.org/10.20517/2394-4722.2020.34
AMA Style
Malentacchi F, Sgromo C, Antonuzzo L, Pillozzi S. Liquid biopsy in endometrial cancer. Journal of Cancer Metastasis and Treatment. 2020; 6: 34. http://dx.doi.org/10.20517/2394-4722.2020.34
Chicago/Turabian Style
Malentacchi, Francesca, Chiara Sgromo, Lorenzo Antonuzzo, Serena Pillozzi. 2020. "Liquid biopsy in endometrial cancer" Journal of Cancer Metastasis and Treatment. 6: 34. http://dx.doi.org/10.20517/2394-4722.2020.34
ACS Style
Malentacchi, F.; Sgromo C.; Antonuzzo L.; Pillozzi S. Liquid biopsy in endometrial cancer. J. Cancer. Metastasis. Treat. 2020, 6, 34. http://dx.doi.org/10.20517/2394-4722.2020.34
About This Article
Special Issue
Copyright
Data & Comments
Data
 Cite This Article 18 clicks
Cite This Article 18 clicks












Comments
Comments must be written in English. Spam, offensive content, impersonation, and private information will not be permitted. If any comment is reported and identified as inappropriate content by OAE staff, the comment will be removed without notice. If you have any queries or need any help, please contact us at support@oaepublish.com.