The use of liquid biopsy in early breast cancer: clinical evidence and future perspectives
Abstract
Liquid biopsy, including both circulating tumor cells and circulating tumor DNA, is gaining momentum as a diagnostic modality adopted in the clinical management of breast cancer. Prospective studies testing several technologies demonstrated clinical validity and, in some cases, achieved the United States Food and Drug Administration approval. The initial testing and clinical application of liquid biopsy focused primarily on the diagnosis, while molecular characterization and monitoring of metastatic disease, with larger data from prospective studies, came in the last two decades. Although its role in metastatic setting is thus widely recognized, the current evidence does not provide support for the routine clinical use of liquid biopsy methods for the earlier stage of this disease. Considering the relevance of early detection, characterization, and management of breast cancer in the early-stage, this clinical setting is the most suitable to increase the chances for effective treatment selection and improved prognosis, and a better understanding of the main application of liquid biopsy tools in the earlier stage of breast cancer is therefore crucial. The aim of this review is to provide an overview of the clinical evidence and subsequent potential applications of liquid biopsy in early breast cancer, identifying the main existing caveats and the possible future scenarios.
Keywords
Introduction
About 276,480 new cases of invasive breast cancer will be diagnosed among American women within 2020[1]. This incidence means breast cancer is the second most common cancer, showing a slightly increasing trend in recent years (by 0.3% per year). Around 6% of these patients will present with metastatic disease at diagnosis, while, among the remaining, approximately 40% will develop a systemic recurrence, despite the surgery and eventual use of systemic and radiation therapy[2]. The 5-year overall survival (OS) rate among patients with localized disease ranges between 86% and 99%. Conversely, only 27% of patients with metastatic breast cancer (mBC) will reach the 5th year after diagnosis[3]. In consideration of these data, it is fundamental that efforts are made to ensure that diagnosis of both primary and potential recurrence is made as early as possible.
The current evidence suggests that mammography remains the only standard of care diagnostic imaging for breast cancer screening (with the exception of high-risk patients, in which case the use of Magnetic Resonance Imaging may be preferred) and, performing a biopsy of the lesion is mandatory to confirm the diagnosis and provide further tumor pathological and molecular characteristics. The standard surveillance methods evaluated for early detection of recurrence of disease are mostly similar to those used for the primary diagnosis. Moreover, some serological markers (such as CA15-3 and CEA) are available for clinical use, although none of these were demonstrated to improve the detection ability of the standard procedures and improve outcome. Tissue biopsies are invasive, time-consuming, quite expensive and for those reasons not often replicable within the same patient. Furthermore, the tissue biopsies appear to be too shortsighted to capture the true spatial and temporal tumor heterogeneity.
The terms liquid biopsy mainly refers to several different diagnostic tests performed on biological fluids (i.e., blood, saliva, urine, etc.) with the aim of investigating the presence of circulating cancer cells (CTCs) or fragments of DNA [circulating tumor DNA (ctDNA)] that can be shed from the tumor. Despite the development and testing of several enrichment techniques to identify, isolate and characterize CTCs in the blood of cancer patients, the only Food and Drug Administration (FDA) approved method is the CellSearchTM (Menarini Silicon Biosystems, Bologna, Italy), that allows the identification and enumeration of CTCs using an immunomagnetic enrichment of epithelial cells by anti-EpCAM antibodies. With this method, the detection of 5 CTCs or more in the peripheral blood of patients with metastatic breast cancer is an independent indicator of worse prognosis[4]. In a retrospective study analyzing data from 18 international centers, the same cutoff identified a group of mBC patients with more aggressive disease irrespective of disease subtype, recurrence site, type and line of therapy suggesting the need to use this test to stratify mBC, Stage IVaggressivevs. Stage IVindolent. In addition, CTCs often bear the same genomic abnormalities detected in metastatic tumor tissue with some additional peculiar phenotypic changes that are responsible for systemic disease spread (epithelial-mesenchymal transition and stem cell markers), or drug resistance (PI3KCA-AKT signaling pathway)[5], suggesting that CTCs may have a key role in the evolution of the disease and can be considered as a sensitive, quantitative and qualitative surrogate of the overall systemic disease.
The median level of circulating DNA is approximately five-fold higher in plasma of breast cancer patients compared to healthy controls[6]. In a sub-cohort of breast cancer patients, ctDNA was detected in both metastatic (90%) and localized disease (55%), and its presence was correlated overall with each stage of the disease and its prognosis[7]. Moreover, ctDNA levels in metastatic breast cancer patients have been associated with tumor burden with a better performance than canonical serological biomarkers[8]. Furthermore, common subtype-specific breast cancer mutations such as phosphatidylinostitol 3-kinase catalytic subunit (PIK3CA) and tumor protein 53 gene (TP53) and resistance genomic abnormalities like Estrogen Receptor 1 (ESR1) and erythroblastic leukemia viral oncogene homolog 2 (ERBB2) can be detected through ctDNA analysis[9,10], proposing it as an effective tool to assess the genomic characteristics and follow the mutant evolution of metastatic breast cancer.
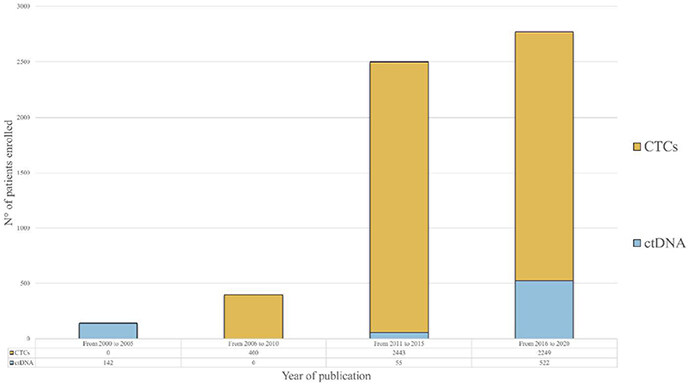
Notwithstanding the well-recognized role of liquid biopsy in the metastatic setting, the importance of these tests for early breast cancer (eBC) patients is still under investigation. However, the increasing number of patients enrolled in investigational trials is evidence of high interest in this matter [Figure 1]. Thus, the aim of this review is to report the current evidence suggesting the potential clinical utility of liquid biopsy for both screening and early diagnosis of recurrence, highlighting the existing limitations and possible future developments.
Figure 1. Histogram representation of patients enrolled in clinical trials assessing the role of CTCs and ctDNA detection in early breast cancer, grouped by year of publication of the results. Showing a relevant rise in the number of enrolled patients in the past 20 years, the chart highlights an increasing interest in liquid biopsy as a tool for the management of early breast cancer patients. CTCs studies have provided a substantial contribution, particularly in the last decade (from 2011 to 2020). A more recent increase in the number of patients enrolled in trials analyzing ctDNA (from 2016 to 2020) suggests a growing interest to investigate the role of ctDNA in this setting. CTCs: circulating tumor cells; ctDNA: circulating tumor DNA
Early diagnosis and baseline characterization
The detection of eBC at pre-clinical stages is the “holy grail” of every diagnostic modality but, also the most challenging. Indeed, the development and clinical testing of highly sensitive diagnostic tools is urgent to ideally reduce BC-related morbidity and mortality, tailor less morbid treatment strategies and eventually improve clinical outcomes. Blood-based detection of circulating cell-free DNA (cfDNA) and the ctDNA fraction holds the great promise to address the need for non-invasive early diagnosis alongside overcoming the shortcomings of tissue biopsies. Moreover, genomic analysis of ctDNA could provide prompt and valuable prognostic information[11]. However, cfDNA and ctDNA levels are significantly low at pre-clinical stages and typically below the level of sensitivity of current clinical testing, affecting the accuracy of ctDNA extraction and analyses through high-throughput sequencing[12]. Indeed, the sensitivity of conventional next generation sequencing (NGS) in detecting DNA alterations is limited to a relatively high fraction of mutant to wild-type DNA (> 1%)[13]. Nonetheless, the conceptual value of ctDNA-based driver gene mutations identification stands in tumor cells clonality, which might in fact determine a high degree of specificity compared to other blood-based biomarkers, endorsing its clinical utility in the diagnostic process.
Beaver et al.[14] prospectively analyzed plasma ctDNA of 29 eBC patients before and after surgery through digital polymerase chain reaction (ddPCR) and compared it with tissue-based Sanger sequencing and ddPCR analyses. PIK3CA mutations were identified in 15 tumor tissues and 13 of them had a matching pre-operatory plasma sample mutation. Of note, in 5 cases PIK3CA mutations were still detectable in post-surgical plasma samples. ddPCR high sensitivity allowed for ctDNA mutation detection with even low levels of tumor burden, providing an initial proof-of-concept to support the feasibility of ctDNA-based analyses in the detection of somatic DNA mutations. Phallen et al.[15] developed the targeted error correction sequencing (TEC-Seq) approach to retrospectively detect cfDNA sequence changes through ultra-sensitive massive genome sequencing. Overall, 58 cancer-related driver genes were analyzed in 200 patients with early-stage colorectal, breast, lung, and ovarian cancer and somatic mutations were detected in 71%, 59%, 59%, and 68% of cases, respectively. Of note, BC ctDNA samples had the lowest mutant allele fraction. High concordance between plasma and tumor-detected alterations was observed and the apparent lack of concordance between some specific alterations, potentially determined by intratumoral heterogeneity, might in fact be overcome by plasma-based analyses.
Recently, a single, multi-analyte blood-test, named CancerSEEK, was developed by Cohen et al.[16] to assess cfDNA mutations and it was applied to 1.005 clinically diagnosed stage I-III cancer patients to screen for eight common cancer types, including eBC. The circulating levels of eight serum protein biomarkers were simultaneously evaluated to overcome the issue of low detectable levels of ctDNA, therefore improving sensitivity. This approach also included machine-learning tools to accurately narrow down the location of a tumor to a small number of anatomic sites. A relatively small yet robust and highly selective 61-amplicon panel was designed through multiplex-polymerase chain reaction to detect ctDNA mutations with an acceptable degree of screening sensitivity and specificity. Median sensitivity was 70% overall and 33% in eBC patients, increasing alongside tumor stage, with 43% for stage I, 73% for stage II, and 78% for stage III cancers. Specificity was higher than 99%, with only 7 out of 812 healthy controls scoring positive and localization of tissue of origin (TOO) was possible in a median of 83% patients. Intriguingly, a significant concordance between ctDNA- and tissue-detected mutations was observed in 90% of cases among all tumors.
Other circulating biomarkers, including exosomes, miRNA, and methylated DNA sequences could be integrated in multi-analyte blood tests to provide complementary early information through gene expression profiling. Proof-of-concept studies have demonstrated that miRNA, packed into tumor-released exosomes, can be detected at higher concentrations compared to ctDNA in early-stage cancer patients. Moreover, their qualitative analyses might be informative both of DNA somatic mutations and epigenetic alterations[17,18]. Indeed, large-scale epigenetic alterations might improve sensitivity in early cancer detection. For instance, the enrichment of methylated cfDNA fragments, which are cancer-specific, could potentially overcome the current technical constraints of mutation-based ctDNA detection methods, mostly determined by the limited amount of recurrent mutations that discriminate tumor and normal cfDNA. Aiming to profile the methylome of small amounts of cfDNA, Shen et al.[19] developed a non-invasive sequencing approach that included cell-free methylated DNA immunoprecipitation and high throughput sequencing (cfMeDIP-seq) for genome-wide bisulfite-free plasma DNA. An optimized Me-DIP-sequencing protocol was first analyzed in a cohort of early-stage pancreatic cancer tissues and healthy controls and DNA methylation profiles were generated through reduced representation bisulfite sequencing (RRBS), demonstrating high analytical sensitivity in detecting cancer-derived DNA. Further analyses suggested the ability of cfMeDIP-seq in detecting tumor-specific methylation events in ctDNA and highlighted that cfMeDIP-seq methylomes could identify active transcriptional networks in tumors or other tissues through plasma cfDNA. CfMeDIP-seq was then explored in a discovery cohort of 189 plasma samples from healthy controls and patients with seven tumor types. Interestingly, this approach demonstrated a high and cost-effective performance in detecting and classifying tumors by recovering ctDNA-associated methylation profiles and it certainly requires further investigation in larger cohorts of eBC patients.
The performance of cfDNA-based targeted methylation sequencing was also evaluated in a prospective case-control sub-study conducted by Liu et al.[20], that assessed the accuracy of a panel of > 10,000 methylation regions to detect and localize a broad range of cancer types in 6,689 participants. In the validation set, sensitivity was 18% among all cancer types, with detection increasing with higher stages, and the specificity was 99.3%. TOO prediction was significantly accurate. This targeted methylation approach allowed deeper sequencing of informative sites compared with whole genome sequencing (WGS).
Cristiano et al.[21] developed a diagnostic tool to investigate genome-wide cfDNA fragmentation patterns of 236 breast, colorectal, lung, ovarian, pancreatic, gastric, or bile duct cancer patients and 245 healthy controls. This study provided further proof-of-concept for plasma-based screening and early cancer detection, as cfDNA altered fragmentation profiles were detected in cancer patients compared to healthy subjects, whose cfDNA rather reflected nucleosomal patterns of white blood cells. In addition, fragmentation profiles were used to confine the TOO to a limited number of sites in 75% of cases.
Besides early diagnosis, plasma-based gene expression profiling allows for the characterization of the genomics, epigenomics, and proteomics of BC patients and ctDNA-based analyses might lead to the identification of targetable mutations at baseline, laying the foundation for very-early precision medicine[22,23].
Granting the value of these preliminary thrilling results, some issues need to be addressed. An accurate blood-based cancer detection test would need a high degree of sensitivity to detect very early-stage disease in a target, asymptomatic, but high-risk population. Notably, most of the studies have evaluated a population of diagnosed but early-stage cancer patients so far. In fact, sensitivity can be improved by deeper sequencing, enhanced error correction methods (including molecular barcoding technology), increased blood amounts and longitudinal evaluation through multiple liquid biopsies. Considering that spontaneous mutation incident rates in healthy subjects may be influenced by multiple non-cancer related biological factors, in addition to sensitivity, specificity is crucial to minimize the risk of high-false positive rates and subsequent unnecessary follow-up tests and to eventually guide the diagnostic workup. Given the impact of early-stage BC diagnosis on cancer-specific survival, the implementation of cfDNA-based cancer detection tools undoubtedly needs further validation in long-term prospective trials to establish its impact on clinical outcomes.
ctDNA dctection at the time of primary diagnosis
The role of ctDNA detection after primary diagnosis was assessed at different timepoints with respect to surgery, neoadjuvant, and adjuvant systemic treatments. In a pioneering prospective trial from Silva et al.[24] (2002), a correlation between the presence of at least 1 out of 6 different chromosomal region’s loss of heterozygosis (LOH), commonly represented among breast carcinomas, or a mutation in TP53 before mastectomy was a predictive factor of disease free survival. Despite the significant results, only 76% of relapsed patients were ctDNA positive. The limited number of alterations explored and the technologies available at that time may explain the low sensitivity.
Taking in consideration the clinical evidence produced in the following years, partly due to the deployment of new technologies, here we address the potential prognostic or predictive role of ctDNA detection before any primary treatment (neoadjuvant, surgical, adjuvant) and the possible implications.
Limited and contrasting evidence is available in support of ctDNA as a predictive biomarker of response to neoadjuvant treatment. In this regard, in a translational ctDNA sub-study of the Neoadjuvant Lapatinib and/or Trastuzumab Treatment Optimisation trial (NeoALTTO)[25], among 69 patients with human epidermal growth factor receptor 2 (HER2) positive+ breast cancer bearing the mutation of TP53 or PIK3CA gene in the primary tumor, the detection of the corresponding alteration in plasma before neoadjuvant treatment was associated with failure to achieve a pathological complete response (pCR). Interestingly, neither detection during week 2 neoadjuvant treatment nor prior surgery were associated with pCR. In this study, ctDNA detection before any treatment was not associated with the time to recurrence (TTR), although the analysis was underpowered to assess the correlation with both TTR and failing to achieve a pCR. Similar results were reported from a prospective study with 55 early breast cancer patients with different subtypes at the diagnosis[26], which demonstrated that ctDNA detection with personalized digital polymerase chain reaction (dPCR) assays of somatic mutations is feasible, but fail to have a prognostic value if detected before neoadjuvant treatment. Conversely, in the same timepoint, a significant association between ctDNA detection and relapse was, instead, found in a bigger prospective cohort of 101 early-stage breast cancer patients [hazard ratio (HR), 5.8; 95% confidence interval (CI): 1.2-27.1; P = 0.01], irrespective of hormone receptor and HER2 status[27]. Due to the conflicting results obtained from these investigations, the prognostic role of ctDNA detection before any treatment remains a matter of debate.
Several other studies addressed the prognostic potential of ctDNA detection before surgery. For instance, despite the modest number of patients recruited on the BRE09-146 phase II clinical trial, Chen et al.[28] were retrospectively capable of identifying a group of triple negative breast cancer (TNBC) patients with residual disease after neoadjuvant treatment with detected ctDNA and a very poor prognosis. Although with high specificity (100%) but very low sensibility (31%), the patients identified by ctDNA detection had a significantly inferior disease-free survival (DFS) if compared with relapsed patients and negative ctDNA (P < 0.0001, median DFS: 4.6 months. vs. Not Reached (NR); HR = 12.6, 95%CI: 3.06-52.2). Considering that only 1 ml of blood was dedicated to this analysis, this study represents the “worst-case scenario” in which the detection of ctDNA identifies an exceptionally high-risk patient population. However, the previous study is not the sole addressing and supporting the potential clinical role of ctDNA in this timepoint. Indeed, to the present day, one of the strongest pieces of evidence is the preplanned analyses from the phase II trial BRE12-158 presented by Radovich et al.[29]. DFS, distant DFS (DDFS), and OS at 24 months of 196 patients with early-stage TNBC, who had residual disease after neoadjuvant treatment, were analyzed. Detection of ctDNA before surgery was significantly associated with an inferior DFS (HR = 2.67, 95%CI: 1.28-87 5.57; P = 0.009), DDFS (median DDFS: 32.5 months vs. Not Reached; HR = 2.99, 95%CI: 1.38-6.48; P = 0.006) and OS (HR = 4.16, 95%CI: 1.66-10.42; P = 0.002). Notably, the distant DFS probability was 56% in ctDNA-positive patients as compared to 81% in ctDNA-negative patients. Taken together, these results indicate that the detection of ctDNA in early-stage TNBC after neoadjuvant treatment is an independent predictor of disease recurrence.
Leveraging the lessons we have learned from these trials, it appears legitimate to further investigate the potential of ctDNA, among the different breast cancer molecular subtypes, to select high risk patients that could benefit from more demanding adjuvant treatment, sparing, in the meanwhile, unnecessary toxicity to those at lower risk of recurrence. Similar strategies are being adopted and investigated in trials enrolling stage II colon cancer. (“The DYNAMIC study- Circulating tumor DNA analysis informing adjuvant chemotherapy in Stage II Colon Cancer”, registration number: ACTRN126150003815830).
ctDNA and detection of minimal residual disease: early diagnosis of recurrence
Patients with potentially curable localized disease will receive surgical treatment and/or eventually radiation and neoadjuvant/adjuvant systemic therapy. The choice of the systemic treatment and the selection of patients that are candidates to receive more aggressive systemic therapies are research fields in constant evolution. Despite the optimal multidisciplinary treatment and the use of appropriate subtype specific systemic therapies, a portion of these patients will relapse as a result of undetected micrometastatic disease. The minimal residual disease (MRD) is the presence of cancer cells that remain in the tissues and/or in the blood after treatment. Considering that the amount of ctDNA, intended as clonal plasma variant allele frequency (VAF), directly correlates to pathological tumor size[30], the detection of ctDNA was hypothesized to be suggestive of MRD. Therefore, several prospective and retrospective trials have addressed the potential of ctDNA to determine those patients who, even if they are considered “disease-free” after surgery, will relapse. In a prospective, multicenter, validation study reported by Garcia-Murillas et al.[27] (2019) after 35.5 months of median follow-up of early breast cancer patients, the detection of ctDNA after surgery was associated with relapse-free survival (HR = 5.8; 95%CI: 1.2-27.1; P = 0.01), with a median lead time of 10.7 months compared with clinical relapse. In this study, the detection of ctDNA trough dPCR assays, based on somatic mutations identified from diagnostic biopsy samples, reached a sensibility of 96% for extracranial distant metastatic relapses. In contrast, brain-only relapses were unlikely to be detected (17%). Similar results were presented by Coombes et al.[31]. In this prospective trial, serial blood draws were performed during a 6-month follow-up, subsequent to surgery and adjuvant treatments, showing a correlation between ctDNA detection in the first post-surgical plasma sample and poorer prognosis (HR = 11.8; 95%CI: 4.3-32.5). In this case, using upfront exome profiling of tumor tissue followed by a personalized targeted multiplex plasma sequencing, ctDNA was detected ahead of metastatic relapse with a median lead-time of 8.9 months. It is worth mentioning that a similar prognostic correlation was shown for ctDNA detection in the successive follow-up plasma samples, giving rise to new question about the potential additional meaning of seriate multiple testing.
Consistent results were reported by Garcia-Murillas et al.[26] (2015), with a median lead time over clinical relapse of 7.9 months. In addition, the authors addressed the above-mentioned question if several repeated sampling after surgery (“mutational tracking”) might outperform the single post-surgical blood draw, showing that repeated samples achieved a higher sensitivity detecting recurrences in estrogen receptor positive (ER+) breast cancer patients. Moreover, in the former study, the alterations highlighted in the ctDNA were more aligned with the alterations found in metastatic biopsy than in primary tumor, supporting the hypothesis that detecting MRD through ctDNA is both a potential prognostic factor and a reliable tool for the molecular characterization of the upcoming metastatic disease.
Despite these promising achievements, sensibility in detecting low concentration of ctDNA in patients apparently “disease-free”, or at least bearing a minimal tumor burden, remains an issue. New technologies are currently being developed and others have been already tested. Among them, in a small exploratory cohort of early breast cancer patients, the targeted digital sequencing (TARDIS) achieved up to a 100-fold improvement beyond the current limit of ctDNA detection[32].
Clinical role of CTCs in early breast cancer
The amount of circulating tumor cells in peripheral blood before the administration of systemic treatment has been proven to be an independent predictor of progression-free survival (PFS) and OS in metastatic breast cancer[4]. However, around 20% early-stage breast cancer patients have CTCs detected in blood[33]. In contrast with the current CTCs positivity cut-off used in metastatic setting, considering the lower detection rate in the earlier stages, a cut-off of 1 or more CTCs was proposed and subsequently adopted in the vast majority of the investigations[34].
The first prospective assessment of the role of CTCs in early breast cancer patients was published in 2006 by Xenidis et al.[35], suggesting that detection of peripheral blood CK-19 mRNA-positive cells in eBC patients were associated with early clinical relapse (P = 0.00001) and disease-related death (P = 0.008). Since then, several big randomized prospective trials preplanned a CTCs analysis with the aim to better evaluate the prognostic and predictive value of CTCs detection in early breast cancer patients.
The main evidences supporting the prognostic value of CTCs detection in the earlier setting of breast cancer were provided by the translational research program of the phase 3 SUCCESS-A trial[36]. Pre- and post-adjuvant treatment blood samples from 2026 patients were evaluated and the presence of CTCs was associated with poor DFS (P < 0.0001), DDFS (P < 0.001), breast cancer-specific survival (P = 0.008), and OS (P = 0.0002). Similar correlations were demonstrated by several other trials, across different subtypes and clinical features[37-41]. Interestingly, the large sized cohort allowed both correlation of CTCs and clinical characteristics (such as node-positivity), and to show an additional negative influence on DFS and OS in case of persistence of CTCs after therapy.
From the same group, a more recent analysis of correlation between CTC counts at the 2-year visit after treatment and outcome provided evidences about follow-up CTCs detection. The presence of CTCs at 2 years after chemotherapy significantly predicted both shorter OS (HR = 4.24, 95%CI: 2.32-7.73; log-rank test, P < 0.001) and shorter DFS (HR = 2.37, 95%CI: 1.57-3.58; log-rank test, P < 0.001) in the univariate analyses. Of note, a subgroup analysis was not able to show a correlation in HER-2 positive breast cancer[42].
The importance of follow-up CTCs detection was more recently assessed in a per-protocol secondary analysis of a double-blinded phase III trial, whose important results were published by Sparano et al.[43] in 2018. Among 353 estrogen receptor positive eBC patients without any evidence of recurrence between 4.5 years and 7.5 years after primary surgery, a positive CTC assay result was associated with a 13.1-fold higher risk of recurrence (hazard ratio point estimate, 13.1; 95%CI: 4.7-36.3), reinforcing the value of CTCs detection during follow-up and the possible future implication in the decisional process on adjuvant endocrine therapy duration of treatment. In the CTCs analysis of REMAGUS 02 trial, Pierga et al.[37] found that circulating tumor cell detection before and/or after neoadjuvant chemotherapy was significantly associated with early metastatic relapse (P = 0.013; median follow-up, 18 months). An updated analysis of this cohort from Bidard et al.[44] confirmed the results with higher statistical significance. However, neither CTC detection before or after neoadjuvant chemotherapy nor changes in CTC count during neoadjuvant chemotherapy was predictive of pathologic complete response.
Patient and treatment selection for neoadjuvant therapy has gained greater momentum in breast cancer since a correlation between tumor response and long-term outcome was observed. However, in contrast with the metastatic setting, the utility of the CTC number change to monitor the efficacy of neoadjuvant treatment has not been demonstrated. In the sub-study of the NeoALTTO phase III trial, a numerically lower pCR rate was observed in patients with detectable CTCs, although no statistically significant association was found[45]. A lack of correlation was highlighted also in the CTCs analyses of the inflammatory breast cancer (IBC) patients enrolled in both BEVERLY 1 and 2 trials, even though the association of achieved pCR and negative baseline CTCs identified a very good prognosis subgroup of HER2+ IBC (95% 3-years DFS)[39,40].
In summary, notwithstanding the potential of CTCs in monitoring treatment response, there is still no solid evidence about their role in driving the treatment intensity as, on the other hand, has been recently suggested in the metastatic setting[46].
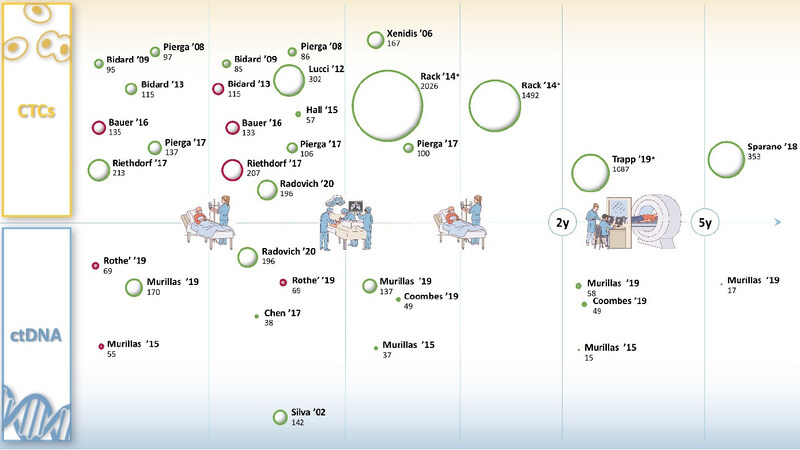
Despite the growing evidence with regards to the potential clinical relevance of pre-treatment detection, several investigations have called into question the value of CTC count after neoadjuvant treatment. The CTCs analysis from the Geparquattro trial[41], after a median follow-up of 67.1 months, showed a correlation between CTCs detection and reduced DFS and OS (P = 0.031 and P = 0.0057) only before neoadjuvant treatment. The lack of prognostic value of post-neoadjuvant detection of CTCs was confirmed in several studies[39,40,47]. However, Hall et al.[48] specifically addressed this question. In a cohort of 57 TNBC, detection of at least 1 CTC after NAT predicted decreased DFS (log-rank P = 0.03, HR = 5.25, 95%CI: 1.34-20.56) and OS (log-rank P = 0.03, HR = 7.04, 95%CI: 1.26-39.35), suggesting a potential different biological behavior across subtypes. For a comprehensive graphic representation of trials assessing the role of CTCs and ctDNA detection as a prognostic biomarker among different timepoints of eBC treatment, see Figure 2.
Figure 2. Bubble representation of the most important clinical trials (name of first author and year of publication) assessing the association between CTCs, ctDNA detection and prognosis. The data are retrieved from original manuscripts and shown in respect of the 5 main management timepoints of early breast cancer (neoadjuvant treatment, surgery, adjuvant treatment, 2 years follow-up, 5 years follow-up). Bubble size represents the number of analyzed patients. Bubble color represents a statistical association between detection and either worse disease-free survival or worse overall survival (green = significant association, red = not significant association). X-axis represents the treatment time frame for which data from the study refers to. Y-axis represents the year of publication: closer to the x axis are the most recent trials. CTCs: circulating tumor cells; ctDNA: circulating tumor DNA. *Bubbles dimension resized by a factor of 3. Figure references: Silva ’02[24], Rothe ’19[25], Murillas ’15[26], Murillas ’19[27], Chen ’17[28], Radovich ’20[29], Coombes ’19[31], Xenidis ’06[35], Rack ’14[36], Pierga ’08[37], Lucci ’12[38], Pierga ’17[40], Trapp ’19[42], Bidard ’09[44], Hall ’15[48], Riethdorf ’17[74], Sparano ’18[76], Bidard ’13[73], Bauer ’16[78]
A combined comparison of the data from the main prospective trials that occurred in the past years has partially answered the above-mentioned questions. In a pooled analysis of 3,173 eBC patients, the presence of CTCs at the time of primary diagnosis was significantly associated with shorter DFS and DDFS, with multivariate HRs of 1.822 (95%CI: 1.470-2.258; P < 0.001) for DFS and 1.888 (95%CI: 1.485-2.401; P < 0.001) for DDFS. The prognostic value of CTCs was significant for all CTC cut-off values ranging from 1 to 20 both for DFS (all P < 0.01) and for DDFS (all P < 0.01) and was confirmed across all subtypes. However, CTC detection did not significantly predict OS in the subgroup of patients with nodal stage N0 disease, showing indeed, higher effect on OS in high risk patients, defined as those with primary tumors larger than 2 cm and lymph node involvement (HR = 2.460; 95%CI: 1.784-3.390; P = 0.001)[49].
Furthermore, in a comprehensive international meta-analysis of 21 trials (the IMENEO study), the number of CTCs detected before NAT administration had no detrimental impact on pathological complete response. Although only a few trials showed that increasing number of CTCs enumerated was correlated with a higher hazard ratio for disease recurrence[29,38], in the former meta-analysis each CTC detected added a quantum of poor prognosis, suggesting that CTCs can be considered as a quantitative marker even in eBC[50].
Caveats and pitfalls of ctDNA in early-stage breast cancer
Notwithstanding the potential of liquid biopsy in the metastatic setting, its deployment for early disease diagnosis, MRD monitoring and characterization still poses multiple challenges[51].
As a matter of fact, ctDNA is directly associated with tumor burden, with consequently lower MAF in early-stage, which is moreover characterized by a significantly low number of ctDNA-detectable aberrations. In addition, low levels of ctDNA could result in low reproducibility, as high pre-analytical and analytical variables could influence the characterization of alterations in situations with a near to the limit of detection MAF[52,53].
Besides tumor burden, low ctDNA levels could also be linked to the differential shedding across metastatic sites. In a study of Serpas et al.[54], knocking-down the DNase1L3 in mice resulted in a change in DNA fragment length, suggesting that different nuclease mechanisms among different tissues could provide more information about the origin of the ctDNA, allowing increased specificity. However, Garcia-Murillas et al.[27] showed that single site relapses were characterized by low, undetectable, ctDNA levels and in particular brain-only relapses were unlikely to be detected with rates of ctDNA detection similar to those of primary brain tumors, probably due to the role of the blood-brain-barrier in hindering ctDNA shedding into the plasma.
All these assumptions translate in the need to develop more sensible techniques, capable of detecting lower and lower ctDNA concentrations.
A first layer of selection could be made by fragment size-based enrichment and selective sequencing that can increase ctDNA detection and could therefore enhance downstream characterizations[55].
Additional mutation-agnostic ctDNA features, such as epigenetics, could potentially increase not only sensitivity, but also specificity[56,57], which is of pivotal importance in the early setting to avoid false negatives with their consequent impact on healthcare costs and patients’ quality of life.
As a matter of fact, the increasingly high sensitivity of sequencing technologies can detect, as a side-effect, somatic mutations that are present across normal tissues. The onset of such confounding somatic mutations may depend on tissue-specific factors, exposure to mutagens as chemotherapy and age[58]. These clones may consequently result in a genetic drift that could potentially cause false-positive results in ctDNA analysis[59].
In particular, the clonal hematopoiesis of indeterminate potential (CHIP) is common with increasing age[60-62]. Garcia-Murillas et al.[27] prospectively assessed the detection of CHIP during MRD monitoring, showing how persistently high levels of truncal mutations such as TP53 or PIK3CA could be detected also in the Buffy coat as a result of CHIP and would otherwise generate false-positive ctDNA results.
Thus, breaking down the barrier of low tumor burden detection through ctDNA analysis, an increasing need to develop highly sensitive and specific techniques has arisen. Several approaches have been previously attempted to combine the two philosophies by applying wide targeted NGS panels or exome sequencing to the primary tumor and consequently screen the resulting mutations on longitudinal ctDNA samples through personalized ddPCR primers[27,32]. Such techniques include BEAMing, SafeSeqS, and TAmSeq[63-65].
Although offering a reasonable balance, these approaches have a potential downside. In fact, adjuvant and neoadjuvant treatments may select new mutant clones that would be therefore missed, with consequently false-negative results.
To overcome the mentioned drawbacks, hybrid capture-based NGS was developed to improve the detection without prior knowledge of primary tumor alterations. In this approach, biotinylated probes, designed against recurrently mutated genomic regions in breast cancer, select relevant DNA sequences for NGS analysis. These technologies allowed the implementation of error-correction strategies (such as integrated digital error suppression) that pushed the ctDNA detection limit to ~ 0.001% (CAPP-Seq[66], TEC-Seq[15]). Commercial assays that utilizes hybrid capture-based NGS technology, as Guardant360®, PlasmaSELECT® and FoundationOne® Liquid, are available for clinical use. However, despite the high self-reported accuracy, specificity and sensitivity, a comparative study of two of these platforms demonstrated a low concordance between the two assays[67].
Caveats and pirfalls of CTCs in early-stage breast cancer
CTC deployment in early-stage breast cancer has encountered several analytical and pre-analytical obstacles. The most challenging limitation encountered by CTC detection in early disease is that CTCs are slightly less represented in blood, compared to physiological cellular components (approximately one cancer cell among a billion normal blood cells, in the metastatic setting). Therefore, considering this setting, even highly sensitive technologies could require a huge amount of blood to detect few CTCs, threatening the clinical feasibility.
Thus, to overcome blood volume limitations, new detection approaches were developed. Inserting a 2 cm long steel wire with a hydrogel functional tip coated with EpCAM antibody, the GILUPI CellCollector is a method that allows to expose the collection tube to approximately 1 L of blood, instead of the canonical 7.5 ml analyzed by CellSearch[68]. However, even if this promising approach has shown a higher CTC capture rate from stage IV and III lung cancer patients compared to the FDA cleared CellSearch, these results have not yet been achieved in a breast cancer patients’ cohort. Furthermore, this technology requires the insertion of a noodle in the cubic vein of the patient for 30 min, resulting to be slightly more invasive than blood draw-based analysis.
CTCs are highly heterogenous and dynamically change their shape and morphology. For instance, downregulation of EpCAM during epithelial-mesenchymal transition (EMT) is a known phenomenon[69].
Accordingly, the inability to select CTCs that underwent to mesenchymal transition represents a further limitation of the EpCAM based CTCs detection approaches. Since the transition of adherent epithelial cells to a migratory mesenchymal state has been implicated in tumor metastasis, as well as in the process in which CTCs acquire greater ability to metastasize[70], new approaches that allow the detection of not-epithelial CTCs are being developed[71].
Among them, is worth of mention the use of carbon nanotubes for adherence-based capture and isolation of CTCs of different phenotypes. This technology, called nanotube-CTCchip, has shown the potential to overcome both selective phenotype detection and size-dependent loss of cells, proposing itself as a versatile platform to capture CTCs of different phenotype[72]. Nevertheless, considering that it has only been tested in a very small cohort of breast cancer patients (7 pts with different clinical staging and 2 healthy controls), this technique is far from clinical deployment.
An additional point of weakness of the literature produced so far in this setting of BC is the arbitrary nature of the positivity cut-off adopted among the different trials. Indeed, in contrast with the well-established prognostic cut-off used in the metastatic setting of 5 CTCs detected in 7.5 ml of blood through the CellSearch, discretionary cut-off of either 1 CTCs, 5 CTCs or both have been used. However, since the number of CTCs appeared to be directly correlated with the prognosis[36] and considering the lower number of CTCs presents in the earlier stages, the lowest cut-off (1 CTCs) has been empirically adopted by most of the investigators.
Conclusion and future directions
Improving the management of early disease patients represents one of the most pressing challenges in breast cancer care. Thus, in order to enhance the chances of early diagnosis, early detection of recurrence and personalized treatment, several liquid biopsy tools have been deployed in this setting in the last 20 years. Among them, CTCs and ctDNA detection and characterization have played a fundamental role, showing a potential prognostic and predictive value in numerous prospective trials.
As a matter of fact, the sensitivity and specificity of the current methods are still suboptimal to candidate ctDNA detection as a tool of early diagnosis. In this context, the assessment of epigenetic alterations (e.g., methylomes analysis) has shown promising preliminary results, demonstrating the potential utility of ctDNA methylation profiles as a basis for non-invasive, sensitive and accurate early tumor detection[19].
Despite the highlighted current analytical and technical limitations of ctDNA detection in early setting, encouraging results are shown by investigations addressing the potential prognostic role of ctDNA detection after primary treatment[27,31], which suggest considering the detection as a good indicator of MRD. Furthermore, the molecular characterization of ctDNA could act as a versatile, dynamic and not invasive tool of disease characterization that might lead clinicians in the process of treatment choice, anticipating or, in a foreseeable future, even partially replacing the tissue biopsy[46].
Combined analysis of ctDNA and CTCs could improve the prognostic value and clinical utility. In fact, under certain circumstances, even with undetectable amount of ctDNA, CTCs can be isolated from the circulation. Indeed, in a cohort of 196 TNBC patients that received a blood draw after NAT (results recently published by Radovich et al.[29]), the combination of ctDNA and CTCs provided additional information to identify patients with worse prognosis, such that the sensitivity to detect recurrences within 24 months went from 79% with ctDNA detection alone to 90% when combined.
These results suggest that these methods have, at least in part, a complementary sensitivity and that the combined use could improve the performances. Another successful combinatorial approach is represented by the PCR-based NGS assay called CancerSEEK, that provided high performance in diagnosis and assessment of primary tumor combining ctDNA detection with different circulating protein biomarkers[16].
However, although the findings provided by several investigations currently suggest a potential clinical impact of ctDNA mutation tracking, clinical utility is still far from being demonstrated, as the highlighted lead time among several studies has not yet been translated in a ctDNA based eBC management that has prospectively shown a significant increase of overall survival.
CTC detection in early-stage breast cancer is associated with prognostic information in patients with locally advanced or high-risk primary breast cancer at different time points in their treatment history[42,47,50]. In particular, the number of CTCs detected before any treatment has been associated with prognosis in several investigations[37,40,44,73,74], suggesting a potential role of CTCs in stratifying patients for neoadjuvant treatment selection. Furthermore, large data analysis including patients with early low-risk luminal breast cancer seem to indicate that CTCs detection could serve as a predictive biomarker for loco regional radiotherapy (XRT)[75]. In that respect, there is currently a plan for a prospective trial to test de-escalating or omitting XRT in this low-risk CTC-negative disease, essentially focusing for the first time on this “indolent” luminal cohort. On the other hand, much of the attention has been focused on patients with detectable CTCs. Studies aimed at using CTC enumeration as selection criteria for additional “adjuvant” therapy have been planned, particularly in hormone-receptor positive disease where CTCs appear associated with late-recurrence and, possibly endocrine resistance[76]. One of such potential, example has been the study testing HER2+ therapies in patients with detectable cells, TREAT-CTC represented an intriguing approach with an ambitious goal but it was stopped for futility due to slow accrual and no indication of benefit[77]. In fact, the study enrolled sixty-three patients with non-metastatic HER2-negative breast cancer and detectable CTCs (> 1/7.5 mL blood) after neoadjuvant chemotherapy and surgery randomized to trastuzumab (Herceptin) or observation. There was no difference in CTCs at week 18, the trial’s primary endpoint, maybe due to CTCs-HER2 definition or true lack of efficacy of the intervention.
In summary, this review showed that in spite of emerging data on the prognostic role of CTCs, the lack of consensus about the “positive” detection cut-off (0, > 1, or > 5), the evaluation timing (at completion of surgery or adjuvant treatment), and the lack of biomarker assessment along with missing clinical utility prospective data may have limited the clinical application of CTC enumeration in early breast cancer. Future studies will evaluate therapeutic strategies for both, de-escalation for CTC-negative disease and, secondary adjuvant for potential late-recurrence in CTC-positive disease. It is likely that combining molecular information such as CTCs, biomarkers, or integration with sensitive and complementary ctDNA technologies may be the key for the practice-changing implementation of liquid biopsy in early disease.
Declarations
Authors’ contributionsMade substantial contributions to conception and design of the work: D’Amico P
Performed acquisition and analysis of data and have drafted the manuscript: D’Amico P, Corvaja C, Gerratana L, Reduzzi C
Revised the manuscript critically and added important intellectual content: Cristofanilli M, Curigliano G
Availability of data and materialsNot applicable.
Financial support and sponsorshipPD is supported by the American-Italian Cancer Foundation Post-Doctoral Research Fellowship, year 2019-2020.
Conflicts of interestAll authors declared that there are no conflicts of interest.
Ethical approval and consent to participateNot applicable.
Consent for publicationNot applicable.
Copyright© The Author(s) 2021.
REFERENCES
1. Society A cancer. Cancer Facts and Figures 2020. Available from: https://www.cancer.org/research/cancer-facts-statistics/all-cancer-facts-figures/cancer-facts-figures-2020.html. [Last accessed on 21 Sep 2020].
2. Colleoni M, Sun Z, Price KN, et al. Annual hazard rates of recurrence for breast cancer during 24 years of follow-up: results from the international breast cancer study group trials I to V. J Clin Oncol 2016;34:927-35.
3. Howlader N, Noone AM, Krapcho M, et al. SEER cancer statistics review, 1975-2016 National Cancer Institute. Bethesda MD. Posted to the SEER web site, April 2019. Available from: https://seer.cancer.gov/csr/1975_2016. [Last accessed on 21 Sep 2020].
4. Cristofanilli M, Budd GT, Ellis MJ, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med 2004;351:781-91.
5. Bredemeier M, Kasimir-Bauer S, Kolberg HC, et al. Comparison of the PI3KCA pathway in circulating tumor cells and corresponding tumor tissue of patients with metastatic breast cancer. Mol Med Rep 2017;15:2957-68.
6. Huang ZH, Li LH, Hua D. Quantitative analysis of plasma circulating DNA at diagnosis and during follow-up of breast cancer patients. Cancer Lett 2006;243:64-70.
7. Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014;6:224ra24.
8. Dawson SJ, Tsui DW, Murtaza M, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med 2013;368:1199-209.
9. Madic J, Kiialainen A, Bidard FC, et al. Circulating tumor DNA and circulating tumor cells in metastatic triple negative breast cancer patients. Int J Cancer 2015;136:2158-65.
10. Rodriguez BJ, Córdoba GD, Aranda AG, et al. Detection of TP53 and PIK3CA Mutations in Circulating Tumor DNA Using Next-Generation Sequencing in the Screening Process for Early Breast Cancer Diagnosis. J Clin Med 2019;8:1183.
11. Alimirzaie S, Bagherzadeh M, Akbari MR. Liquid biopsy in breast cancer: A comprehensive review. Clin Genet 2019;95:643-60.
12. Chen KZ, Lou F, Yang F, et al. Circulating tumor DNA detection in early-stage non-small cell lung cancer patients by targeted sequencing. Sci Rep 2016;6:31985.
13. Zhang XF, Ju SQ, Wang XD, Cong H. Advances in liquid biopsy using circulating tumor cells and circulating cell-free tumor DNA for detection and monitoring of breast cancer. Clin Exp Med 2019;19:271-9.
14. Beaver JA, Jelovac D, Balukrishna S, et al. Detection of cancer DNA in plasma of early stage breast cancer patients. Clin Cancer Res 2014;20:2643-50.
15. Phallen J, Sausen M, Adleff V, et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci Transl Med 2017;9:eaan2415.
16. Cohen JD, Li L, Wang Y, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018;359:926-30.
17. Salido-Guadarrama I, Romero-Cordoba S, Peralta-Zaragoza O, Hidalgo-Miranda A, Rodríguez-Dorantes M. MicroRNAs transported by exosomes in body fluids as mediators of intercellular communication in cancer. Onco Targets Ther 2014;7:1327-38.
18. Gold B, Cankovic M, Furtado LV, Meier F, Gocke CD. Do circulating tumor cells, exosomes, and circulating tumor nucleic acids have clinical utility? A report of the association for molecular pathology. J Mol Diagn 2015;17:209-24.
19. Shen SY, Singhania R, Fehringer G, et al. Sensitive tumour detection and classification using plasma cell-free DNA methylomes. Nature 2018;563:579-83.
20. Liu M, Oxnard G, Klein E, et al. Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Annals of Oncology 2020;31:745-59.
21. Cristiano S, Leal A, Phallen J, et al. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature 2019;570:385-9.
22. Gerratana L, Davis AA, Shah AN, Lin C, Corvaja C, Cristofanilli M. Emerging Role of Genomics and Cell-Free DNA in Breast Cancer. Curr Treat Options Oncol 2019;20:68.
23. Koboldt DC, Fulton RS, McLellan MD, et al. Comprehensive molecular portraits of human breast tumors. Nature 2013;490:61-70.
24. Silva JM, Silva J, Sanchez A, et al. Tumor DNA in plasma at diagnosis of breast cancer patients is a valuable predictor of disease-free survival. Clin Cancer Res 2002;8:3761-6.
25. Rothé F, Silva MJ, Venet D, et al. Circulating tumor DNA in HER2-amplified breast cancer: a translational research substudy of the NeoALTTO phase III trial. Clin Cancer Res 2019;25:3581-8.
26. Garcia-Murillas I, Schiavon G, Weigelt B, et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Transl Med 2015;7:302ra133.
27. Garcia-Murillas I, Chopra N, Comino-Méndez I, et al. Assessment of molecular relapse detection in early-stage breast cancer. JAMA Oncol 2019;5:1473-8.
28. Chen YH, Hancock BA, Solzak JP, et al. Next-generation sequencing of circulating tumor DNA to predict recurrence in triple-negative breast cancer patients with residual disease after neoadjuvant chemotherapy. NPJ Breast Cancer 2017;3:24.
29. Radovich M, Jiang G, Hancock BA, et al. Association of circulating tumor DNA and circulating tumor cells after neoadjuvant chemotherapy with disease recurrence in patients with triple-negative breast cancer: preplanned secondary analysis of the BRE12-158 randomized clinical trial. JAMA Oncol 2020;6:1410-5.
30. Swanton C, Jamal-Hanjani M, Abbosh C, et al. Phylogenetic ctDNA analysis depicts early stage lung cancer evolution. Nature 2017;545:446-51.
31. Coombes RC, Page K, Salari R, et al. Personalized detection of circulating tumor DNA antedates breast cancer metastatic recurrence. Clin Cancer Res 2019;25:4255-63.
32. McDonald BR, Contente-Cuomo T, Sammut SJ, et al. Detection of residual disease after neoadjuvant therapy in breast cancer using personalized circulating tumor DNA analysis. Sci Transl Med 2019;7392:1-14.
33. Krishnamurthy S, Cristofanilli M, Singh B, et al. Detection of minimal residual disease in blood and bone marrow in early stage breast cancer. Cancer 2010;116:3330-7.
34. Tibbe AG, Miller MC, Terstappen LW. Statistical considerations for enumeration of circulating tumor cells. Cytometry A 2007;71:154-62.
35. Xenidis N, Perraki M, Kafousi M, et al. Predictive and prognostic value of peripheral blood cytokeratin-19 mRNA-positive cells detected by real-time polymerase chain reaction in node-negative breast cancer patients. J Clin Oncol 2006;24:3756-62.
36. Rack B, Schindlbeck C, Jückstock J, et al. Circulating tumor cells predict survival in early average-to-high risk breast cancer patients. J Natl Cancer Inst 2014;106:dju066.
37. Pierga JY, Bidard FC, Mathiot C, et al. Circulating tumor cell detection predicts early metastatic relapse after neoadjuvant chemotherapy in large operable and locally advanced breast cancer in a phase II randomized trial. Clin Cancer Res 2008;14:7004-10.
38. Lucci A, Hall CS, Lodhi AK, et al. Circulating tumour cells in non-metastatic breast cancer: a prospective study. The Lancet Oncology 2012;13:688-95.
39. Pierga JY, Petit T, Lévy C, et al. Pathological response and circulating tumor cell count identifies treated HER2+ inflammatory breast cancer patients with excellent prognosis: BEVERLY-2 survival data. Clin Cancer Res 2015;21:1298-304.
40. Pierga JY, Bidard FC, Autret A, et al. Circulating tumour cells and pathological complete response: independent prognostic factors in inflammatory breast cancer in a pooled analysis of two multicentre phase II trials (BEVERLY-1 and -2) of neoadjuvant chemotherapy combined with bevacizumab. Ann Oncol 2017;28:103-9.
41. Riethdorf S, Müller V, Zhang L, et al. Detection and HER2 expression of circulating tumor cells: prospective monitoring in breast cancer patients treated in the neoadjuvant GeparQuattro trial. Clin Cancer Res 2010;16:2634-45.
42. Trapp E, Janni W, Schindlbeck C, et al. Presence of circulating tumor cells in high-risk early breast cancer during follow-up and prognosis. J Natl Cancer Inst 2019;111:380-7.
43. Sparano JA, Henry NL. Surveillance after treatment of localized breast cancer: time for reappraisal? J Natl Cancer Inst 2019;111:339-41.
44. Bidard FC, Mathiot C, Delaloge S, et al. Single circulating tumor cell detection and overall survival in nonmetastatic breast cancer. Ann Oncol 2010;21:729-33.
45. Azim HA Jr, Rothé F, Aura CM, et al. Circulating tumor cells and response to neoadjuvant paclitaxel and HER2-targeted therapy: a sub-study from the NeoALTTO phase III trial. Breast 2013;22:1060-5.
46. Bidard FC, Jacot W, Kiavue N, et al. Efficacy of circulating tumor cell count-driven vs clinician-driven first-line therapy choice in hormone receptor-positive, ERBB2-negative metastatic breast cancer: the STIC CTC randomized clinical trial. JAMA Oncol 2020;5:e205660.
47. Bauer ECA, Schochter F, Widschwendter P, et al. Prevalence of circulating tumor cells in early breast cancer patients 2 and 5 years after adjuvant treatment. Breast Cancer Res Treat 2018;171:571-80.
48. Hall C, Karhade M, Laubacher B, et al. Circulating tumor cells after neoadjuvant chemotherapy in stage I-III triple-negative breast cancer. Ann Surg Oncol 2015;22:S552-8.
49. Janni WJ, Rack B, Terstappen LW, et al. Pooled analysis of the prognostic relevance of circulating tumor cells in primary breast cancer. Clin Cancer Res 2016;22:2583-93.
50. Bidard FC, Michiels S, Riethdorf S, et al. Circulating tumor cells in breast cancer patients treated by neoadjuvant chemotherapy: a meta-analysis. J Natl Cancer Inst 2018;110:560-7.
51. Davis AA, Jacob S, Gerratana L, et al. Abstract P5-01-08: landscape of circulating tumor DNA (ctDNA) in metastatic breast cancer. Cancer Res 2020;80:P5-01-08.
52. Paweletz CP, Lau CJ, Oxnard GR. Does testing error underlie liquid biopsy discordance? JCO Precision Oncology 2019;3:1-3.
53. Gerratana L, Zhang Q, Shah AN, et al. Performance of a novel next generation sequencing circulating tumor DNA (ctDNA) platform for the evaluation of samples from patients with metastatic breast cancer (MBC). Crit Rev Oncol Hematol 2020;145:102856.
54. Serpas L, Chan RWY, Jiang P, et al. Dnase1l3 deletion causes aberrations in length and end-motif frequencies in plasma DNA. Proc Natl Acad Sci U S A 2019;116:641-9.
55. Mouliere F, Chandrananda D, Piskorz AM, et al. Enhanced detection of circulating tumor DNA by fragment size analysis. Sci Transl Med 2018;10:eaat4921.
56. Liu L, Toung JM, Jassowicz AF, et al. Targeted methylation sequencing of plasma cell-free DNA for cancer detection and classification. Ann Oncol 2018;29:1445-53.
57. Gerratana L, Basile D, Franzoni A, et al. Plasma-based longitudinal evaluation of ESR1 epigenetic status in hormone receptor-positive HER2-negative metastatic breast cancer. Front Oncol 2020;10:550185.
58. Maltoni R, Casadio V, Ravaioli S, et al. Cell-free DNA detected by “liquid biopsy” as a potential prognostic biomarker in early breast cancer. Oncotarget 2017;8:16642-9.
59. Yizhak K, Aguet F, Kim J, et al. RNA sequence analysis reveals macroscopic somatic clonal expansion across normal tissues expansion across normal tissues. Science 2019;364:eaaw0726.
60. Hu Y, Ulrich BC, Supplee J, et al. False-positive plasma genotyping due to clonal hematopoiesis. Clin Cancer Res 2018;24:4437-43.
61. Swanton C, Venn O, Aravanis A, et al. Prevalence of clonal hematopoiesis of indeterminate potential (CHIP) measured by an ultra-sensitive sequencing assay: exploratory analysis of the Circulating Cancer Genome Atlas (CCGA) study. J Clin Oncol 2018;36:12003.
62. Heuser M, Thol F, Ganser A. Clonal hematopoiesis of indeterminate potential. Dtsch Arztebl Int 2016;113:317-22.
63. Diehl F, Li M, He YP, Kinzler KW, Vogelstein B, Dressman D. BEAMing: single-molecule PCR on microparticles in water-in-oil emulsions. Nat Methods 2006;3:551-9.
64. Kinde I, Wu J, Papadopoulos N, Kinzler KW, Vogelstein B. Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci U S A 2011;108:9530-5.
65. Forshew T, Murtaza M, Parkinson C, et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med 2012;4:136ra68.
66. Newman AM, Bratman SV, To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 2014;20:548-54.
67. Kim AS, Bartley AN, Bridge JA, et al. Comparison of laboratory-developed tests and FDA-approved assays for BRAF, EGFR, and KRAS testing. JAMA Oncol 2018;4:838-41.
68. Gorges TM, Penkalla N, Schalk T, et al. Enumeration and Molecular Characterization of Tumor Cells in Lung Cancer Patients Using a Novel In vivo Device for Capturing Circulating Tumor Cells. Clin Cancer Res 2016;22:2197-206.
69. Hyun KA, Koo GB, Han H, et al. Epithelial-to-mesenchymal transition leads to loss of EpCAM and different physical properties in circulating tumor cells from metastatic breast cancer. Oncotarget 2016;7:24677-87.
70. Yu M, Bardia A, Wittner BS, et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 2013;339:580-4.
71. Raimondi C, Nicolazzo C, Gradilone A. Circulating tumor cells isolation: the “post-EpCAM era”. Chin J Cancer Res 2015;27:461-70.
72. Loeian MS, Mehdi Aghaei S, Farhadi F, et al. Liquid biopsy using the nanotube-CTC-chip: capture of invasive CTCs with high purity using preferential adherence in breast cancer patients. Lab Chip 2019;19:1899-915.
73. Bidard FC, Belin L, Delaloge S, et al. Time-dependent prognostic impact of circulating tumor cells detection in non-metastatic breast cancer: 70-month analysis of the REMAGUS02 study. Int J Breast Cancer 2013;2013:130470.
74. Riethdorf S, Müller V, Loibl S, et al. Prognostic impact of circulating tumor cells for breast cancer patients treated in the neoadjuvant “Geparquattro” trial. Clin Cancer Res 2017;23:5384-93.
75. Goodman CR, Seagle BL, Friedl TWP, et al. Association of circulating tumor cell status with benefit of radiotherapy and survival in early-stage breast cancer. JAMA Oncol 2018;4:e180163.
76. Sparano J, O’Neill A, Alpaugh K, et al. Association of circulating tumor cells with late recurrence of estrogen receptor-positive breast cancer: a secondary analysis of a randomized clinical trial. JAMA Oncol 2018;4:1700-6.
77. Ignatiadis M, Litière S, Rothe F, et al. Trastuzumab versus observation for HER2 nonamplified early breast cancer with circulating tumor cells (EORTC 90091-10093, BIG 1-12, Treat CTC): a randomized phase II trial. Ann Oncol 2018;29:1777-83.
Cite This Article
Export citation file: BibTeX | RIS
OAE Style
D’Amico P, Corvaja C, Gerratana L, Reduzzi C, Curigliano G, Cristofanilli M. The use of liquid biopsy in early breast cancer: clinical evidence and future perspectives. J Cancer Metastasis Treat 2021;7:3. http://dx.doi.org/10.20517/2394-4722.2020.93
AMA Style
D’Amico P, Corvaja C, Gerratana L, Reduzzi C, Curigliano G, Cristofanilli M. The use of liquid biopsy in early breast cancer: clinical evidence and future perspectives. Journal of Cancer Metastasis and Treatment. 2021; 7: 3. http://dx.doi.org/10.20517/2394-4722.2020.93
Chicago/Turabian Style
D’Amico, Paolo, Carla Corvaja, Lorenzo Gerratana, Carolina Reduzzi, Giuseppe Curigliano, Massimo Cristofanilli. 2021. "The use of liquid biopsy in early breast cancer: clinical evidence and future perspectives" Journal of Cancer Metastasis and Treatment. 7: 3. http://dx.doi.org/10.20517/2394-4722.2020.93
ACS Style
D’Amico, P.; Corvaja C.; Gerratana L.; Reduzzi C.; Curigliano G.; Cristofanilli M. The use of liquid biopsy in early breast cancer: clinical evidence and future perspectives. J. Cancer. Metastasis. Treat. 2021, 7, 3. http://dx.doi.org/10.20517/2394-4722.2020.93
About This Article
Special Issue
Copyright
Data & Comments
Data
 Cite This Article 99 clicks
Cite This Article 99 clicks











Comments
Comments must be written in English. Spam, offensive content, impersonation, and private information will not be permitted. If any comment is reported and identified as inappropriate content by OAE staff, the comment will be removed without notice. If you have any queries or need any help, please contact us at support@oaepublish.com.