Utility of circulating tumor DNA for predicting prognosis in the management of resectable pancreatic cancer
Abstract
Background: The measurement of circulating tumor DNA (ctDNA) has been studied in several malignancies, including metastatic pancreatic cancer, but less is known about its utility in monitoring treatment response and recurrence in resectable pancreatic cancer. Methods: We conducted a systematic review of the literature examining the association of ctDNA with overall survival (OS) and disease-free survival. Results: Five articles met our exclusion criteria. Baseline and/or postoperative ctDNA was found to be associated with decreased OS and recurrence-free survival. Discussion: ctDNA has the potential to be used as a prognostic biomarker and to guide therapy in resectable pancreatic cancer.
Keywords
INTRODUCTION
Pancreatic ductal adenocarcinoma (PDAC) is the third leading cause of cancer-related death in the United States, with a 5-year survival rate of approximately 10%[1,2]. While surgical resection is the only curative treatment for pancreatic cancer, up to 80% of diagnosed patients are not candidates for resection due to aggressive spread and advanced-stage diagnosis[3]. For the 20% who undergo curative resection, recurrence occurs in up to 80% of cases[4], likely due to micro-metastatic disease present at the time of diagnosis[5,6].
Current methods for diagnosis, treatment planning, and surveillance of pancreatic cancer are insufficient. The only serum biomarker recommended by the National Comprehensive Cancer Network guidelines is serum carbohydrate antigen 19-9 (CA 19-9), which can be used as an adjunct to guide treatment plans for patients, informing the diagnosis, resectability, response to therapy, recurrence, and overall prognosis of the disease[7]. However, testing CA 19-9 levels can result in false positives due to the presence of certain medical conditions[8-11], and false negatives in those who are Lewis genotype negative and do not express CA 19-9[12]. More accurate biomarkers like circulating tumor DNA (ctDNA) could be valuable for recurrence risk stratification, therapeutic decision making, and detecting and monitoring disease burden in pancreatic cancer.
In 1946, Mandel and Metais described DNA in noncellular blood, which was referred to as cell-free DNA (cfDNA)[13]. Further studies demonstrated that patients with cancer had higher circulating levels of cfDNA[14]. In patients with cancer, DNA released from tumor cells, ctDNA, makes up a fraction of the overall cfDNA. Detection of tumor-specific ctDNA is a highly specialized process[15,16]. There are two main methods of ctDNA detection and quantification, digital droplet polymerase chain reaction (ddPCR), and next-generation sequencing (NGS). ddPCR involves the detection of preselected genes (usually KRAS for pancreatic cancer) in each sample. Compared to traditional PCR, a single sample is partitioned into tens of thousands of nanoliter-sized droplets, allowing the measurement of thousands of independent amplification events in a single sample[17]. However, for multiple gene targets, the use of ddPCR can be cumbersome, which is where NGS demonstrates its utility. With NGS, mutated gene sequences do not need to be preselected. NGS platforms perform sequencing of millions of small fragments of DNA in parallel and compare them against a human reference genome. This allows NGS to sequence entire exomes or genomes and identify novel sequences at an extremely high resolution. However, because the sequencing is more extensive, it is more time-consuming and expensive than ddPCR, especially for low numbers of desired gene targets[18]. Though ctDNA levels often parallel tumor burden within individual patients, variability has been observed in patients with the same form of cancer, possibly reflecting individual tumor biology and rate of cell turnover[19,20]. Unlike currently utilized serological biomarkers such as carcinoembryonic antigen (CEA) and cancer antigen 125 (CA-125), ctDNA has a short half-life (approximately 1 h)[11,21]. Because of this, ctDNA may be used to more accurately monitor the time-related response of an individual patient to therapy[22]. There is also potential for improved surveillance after treatment, where, in some studies, increased ctDNA can precede radiographic disease progression by months[22-25].
ctDNA has gained traction in gastrointestinal malignancies[26-28]; however, its use in pancreatic cancer remains investigational. Measuring ctDNA in pancreatic cancer has shown promise in monitoring locally advanced and metastatic tumors[29-31], but less is known about how ctDNA can inform the management of localized, non-metastatic pancreatic cancer. This report reviews the literature on the prognostic value of ctDNA in resectable pancreatic cancer and the potential value of assessment during treatment.
METHODS
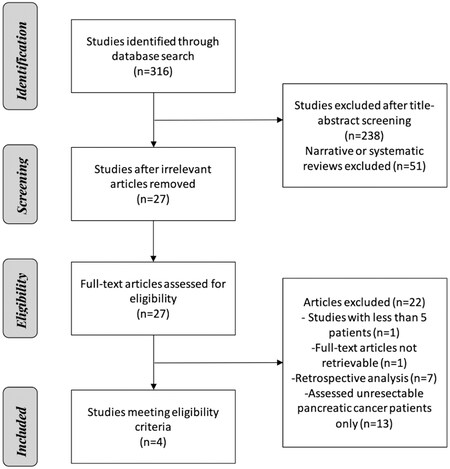
A computerized search was completed up to July 2020 on PubMed with no date restrictions. The search strategy included (pancreatic cancer OR PDAC OR pancreatic ductal adenocarcinoma) AND (cell-free DNA OR cfDNA OR circulating tumor DNA OR ctDNA) AND (resect OR local OR surgical OR operative OR early-stage OR early). This search generated 316 articles.
Titles and abstracts were screened by three authors, and all articles not pertaining to ctDNA in patients with pancreatic cancer were excluded, as were reviews and meta-analyses. Disagreements between the first two authors were decided by a third author. A full-text analysis of the remaining articles was then completed with a focus on overall survival (OS) and disease-free survival. Articles with a study population containing only unresectable or metastatic pancreatic cancer cases or a sample size of fewer than five patients were excluded, as were retrospective studies. The study selection process resulted in the collection of four studies meeting our eligibility criteria, including prospective nature and pancreatic cancer patients that had undergone surgical resection [Figure 1].
RESULTS
ctDNA may be useful as a prognostic biomarker in patients with resectable pancreatic cancer. Baseline and/or postoperative ctDNA has been associated with decreased OS and recurrence-free survival (RFS) in several prospective studies, as summarized in Table 1.
Prospective studies of ctDNA in resectable pancreatic cancer
| Year | Journal | Author | No. of subjects | No. underwent resection | ctDNA detected preoperatively (%) | ctDNA detected postoperatively (%) | Stage | Target candidate | Detection method | Detection time point | Median follow-up (months) | Median overall survival (months) | Median recurrence free survival (months) | ||
| ctDNA+ | ctDNA- | ctDNA+ | ctDNA- | ||||||||||||
| 2019 | Ann Oncol | Lee et | 112 | 81 | 62 (23/37) | 37 (13/35) | Resectable | KRAS (Codons 12,13,61) | PCR-based SafeSeqS | Preoperatively, postoperatively | 38.4 | Preop 13.6; Postop 10.6 | Preop NR; Postop NR | Preop 10.3; Postop 5.4 | Preop NR; Postop 17.1 |
| 2019 | Clin Cancer Res | Groot et al.[34] | 59 | 59 | 49 (29/59) | 26 (11/41) | Resectable | KRAS (G12D, G12V, G12R, Q61H) | ddPCR | Preoperatively, postoperatively | 16 | Preop 14; Postop 17 | Preop NR; Postop NR | Preop 8; Postop 5 | Preop 19; Postop 15 |
| 2018 | Clin Chem | Kim et al.[35] | 106 | 41 | 85 (35/41) | NA | Resectable, locally advanced, metastatic | KRAS (G12A, G12C, G12D, G12R, G12S, G12V, G13D) | ddPCR | At Diagnosis | 10.3 | High MC and MA : ~7 Low MC and MA: NR | High MC and MA:NR Low MC and MA: NR* | ||
| 2017 | Clin Cancer Res | Pietrasz et al.[33] | 135 | 31 | NA | 19 (6/31) | All | KRAS (G12D, G12V, G12R) | dPCR | Postoperatively | 33.3 | 19.3 | 32.2 | 4.6 | 17.6 |
In a prospective multi-institutional study, Lee et al. evaluated 81 pancreatic cancer patients undergoing curative resection[32]. Plasma was tested for ctDNA pre- and postoperatively. Preoperative ctDNA detection (ctDNA positive disease [ctDNA+]) was associated with inferior RFS compared with ctDNA negative (ctDNA-) patients who had undetectable ctDNA levels. (ctDNA+: 10.3 months vs. ctDNA-: median not reached [NR]; HR 4.1; 95%CI: 1.8-9.0, P = 0.002]. OS was also significantly lower [OS 13.6 months vs. NR (HR 4.1, 95%CI: 1.6-10.5; P = 0.015)]. Based on multivariate analysis adjusted for preoperative and pathologic factors, preoperative detection of ctDNA had a statistically significant association with recurrence (HR 4.1; 95%CI: 1.4-12.1; P = 0.018) and OS (HR 4.1; 95%CI: 1.0-16.6; P = 0.049).
After curative intent surgical resection, patients with persistent ctDNA detection postoperatively had a shorter interval of recurrence compared to those who remained undetectable (5.4 vs. 17.1 months; HR 5.4, CI 1.9-15.2; P < 0.001) and shorter OS (10.6 months vs. NR; HR 4.0, CI 1.2-13.6; P = 0.003). Of note, in the 13 patients who were ctDNA+ postoperatively, 100% had cancer recurrence during the median follow-up period of 38.4 months from the time of diagnosis. In comparison, in the 22 patients who were ctDNA- postoperatively, 10 (45%) had disease recurrence during the study period. Detectable ctDNA following curative resection was independently associated with recurrence, with a 100% positive predictive value, with a specificity and sensitivity of 100% and 57%, respectively. The authors concluded that postoperative ctDNA detection has the best prognostic value in predicting recurrence compared with preoperative ctDNA and other clinical and pathologic factors[32].
In a similar study, Pietrasz et al. found postoperative ctDNA detection was prognostic of OS and RFS[33]. In the subgroup of 31 patients who underwent curative intent resection for pancreatic cancer, patients with detectable ctDNA postoperatively had a median OS of 19.3 months, compared to 32.3 months for patients with undetectable ctDNA (P = 0.027). Similarly, ctDNA+ patients had an RFS of 4.6 months vs. 17.6 months in ctDNA- patients. Of note, in the subgroup of patients who underwent resection, preoperative ctDNA studies were not collected[33].
In a prospective study of 59 patients with resectable PDAC, Groot and colleagues collected preoperative and longitudinal postoperative plasma samples for the detection of ctDNA[34]. Subjects had a median follow-up of 16 months from the time of surgical resection. Patients with detectable preoperative ctDNA had a decreased median RFS and incidence of recurrence (8 months; 26 out of 29 [90%] recurred), compared with ctDNA- patients (19 months; 15 out of 30 [50%] recurred; P < 0.001). Similarly, preoperatively ctDNA+ patients had a lower median OS of 14 months (18 out of 29 [62%] died), while median OS was not reached in the ctDNA- group (8 out of 30 [27%] died; P < 0.001). In multivariable Cox regression models adjusted for preoperative factors, preoperative ctDNA detection was an independent predictor of RFS (HR = 2.67; P = 0.011) and OS (HR = 2.37; P = 0.048).
Postoperative ctDNA+ status was also associated with poor RFS and OS. In the 11 patients who were ctDNA+ postoperatively (median follow-up for this sub-group was 15 months from the time of surgery), the median RFS was five months (95%CI: 3-8) compared with 15 months (95%CI: 8-22) in the 30 ctDNA- patients (P < 0.001). Similarly, the median OS was 17 months in ctDNA+ patients (95%CI: 10-24) vs. not reached in the ctDNA- group (P = 0.011). Notably, 24 of the 59 patients in this study underwent neoadjuvant chemotherapy prior to surgical resection. Those patients were significantly less likely to be ctDNA+ in their preoperative sample compared to chemotherapy naïve patients (21% vs. 69%; P < 0.001). The five patients who underwent neoadjuvant chemotherapy and were ctDNA+ preoperatively had poor outcomes; all five patients had disease recurrence with a median RFS of five months, and 2 out of 5 patients died after a 12-month median follow-up period. There were no statistically significant differences in ctDNA positivity based on chemotherapy regimen or duration of treatment[34].
Kim et al. examined the association with concentration and fractional abundance of ctDNA KRAS mutations at the time of diagnosis with outcomes in a study of 106 patients with PDAC[35]. Of 41 patients with resectable tumors, 35 (85%) were ctDNA+ at diagnosis. The KRAS mutation concentration of 0.165 copies/μL and a median fractional abundance of 0.415% were utilized as the cutoff to dichotomize “high” vs. “low” in their dataset. Among those with resectable disease, patients with high KRAS mutation concentration experienced significantly lower median progression-free survival (PFS, defined as the interval from the day of diagnosis to the day of progression or death) compared to those in the low group (7 months vs. not reached, P = 0.016); but no statistically significant difference in OS (8 months vs. not reached
DISCUSSION
This systematic review highlights the utility of ctDNA as a prognostic indicator and biomarker in resectable or potentially resectable pancreatic cancer in the pre- and postoperative setting. As high-quality prospective studies continue to deliver additional data on the utility of this technology in pancreatic cancer, ctDNA has the potential to become a standard of care biomarker, which may help to stratify risk for recurrence and inform therapy decisions. Detection of ctDNA is accomplished within the normal clinical workflow. This blood test can be added to routine blood work during pre- and postoperative visits.
As its detection may identify patients who are at higher risk for recurrence, ctDNA is a potential tool for neoadjuvant, surgical, and adjuvant therapy decision making. In Lee et al.’s study, all 13 patients with postoperatively detectable ctDNA had disease recurrence, despite all receiving adjuvant therapy[32], suggesting that current adjuvant therapy regimens may not be as beneficial in patients who are ctDNA+ postoperatively, as they may be at higher risk for recurrence. However, it is still unclear if clearance of ctDNA during adjuvant therapy portends a better outcome. Persistent ctDNA detectability during adjuvant therapy may indicate the need for an alternative therapy. In line with this, 83% of patients in this study with detectable ctDNA at diagnosis had disease recurrence, despite 81% undergoing standard adjuvant therapy[32], suggesting that preoperative ctDNA detection may be a factor in determining who may benefit from neoadjuvant therapy, rather than proceeding directly to surgery.
Groot and colleagues demonstrated that patients who underwent neoadjuvant therapy were significantly less likely to have ctDNA detected preoperatively (21% vs. 69% P < 0.001)[34]. Notably, the five patients with persistent ctDNA after neoadjuvant therapy had a particularly poor prognosis, with 100% recurrence and a median RFS of five months; two of whom had an overall survival shorter than 12 months[34]. Although this is a small sample size, these results favor avoiding surgery or prolonging therapy in patients with persistent ctDNA after neoadjuvant chemotherapy. Current prospective clinical trials are underway to better define the utility and predictive value of ctDNA to monitor for treatment response in pancreatic cancer. For example, Chawla and colleagues are assessing ctDNA detectability in a phase II prospective observational trial in patients undergoing neoadjuvant chemotherapy for potentially resectable pancreatic cancer. The study aims to determine if clearance of ctDNA during treatment correlates with traditionally accepted treatment response criteria like serum CA 19-9, histologic regression grade, and histologic margin assessment. (NCT04616131). Several other studies examining longitudinal changes in ctDNA levels in response to treatment modalities are ongoing. Heyd and colleagues are conducting a 3-year longitudinal study of ctDNA in patients with resectable and potentially resectable pancreatic cancer to determine its utility in monitoring treatment response and predicting RFS. Serum samples are analyzed at diagnosis, after neoadjuvant chemotherapy, preoperatively, at the time of surgery, 1 month postoperatively, and 1 month after the last round of adjuvant chemotherapy (NCT02818907). In a similar study, Woo and colleagues aim to generate a ctDNA gene panel and collect longitudinal serum samples to verify the applicability of predictive serum-based biomarkers for pancreatic cancer. Their patient population includes patients with both resectable and metastatic pancreatic cancers, from whom they are collecting serum samples every 1-3 months for 24 months (NCT04241367).
Though ctDNA currently shows promise as a prognostic indicator and a tool to measure treatment response, it also has potential as a diagnostic biomarker and screening test, especially when used alongside other serum biomarkers and imaging. CA 19-9 has a sensitivity of 58%-87% and a specificity of 70%-80%, which is inadequate in diagnosing early-stage tumors[36-38]. Dabritz and colleagues found that a combination of testing CA 19-9 levels and KRAS ctDNA led to a sensitivity of 91% in detecting pancreatic cancer[36]. Similarly, Wang et al. found that testing CA 19-9 in combination with ctDNA led to an increased sensitivity of 82% with an 81% specificity in their study of 100 pancreatic cancer patients[39]. ctDNA, as a screening test for pancreatic cancer, may have further utility in high-risk individuals. In a current prospective trial, patients with a strong family history of pancreatic cancer are evaluated with MRIs and plasma ctDNA assessment every 6 months for 3 years to correlate biomarker changes with radiological abnormalities as they arise on imaging (NCT03250078). Ongoing prospective studies evaluating ctDNA in pancreatic cancer are summarized in Table 2.
Current trials exploring utility of ctDNA in pancreatic cancer
| Trial number | Trial name | ctDNA focus and aims | Study type | Estimated completion | Recruitment status |
| NCT04616131 | Evaluating the response to neoadjuvant chemotherapy with circulating tumor DNA in pancreatic cancer | Detection of ctDNA in patients with resectable pancreatic cancer before, during and after treatment and comparison with Ca-19-9, histologic regression, and margin assessment | Prospective observational cohort | October 2021 | Recruiting |
| NCT02818907 | Evaluation of survival prognostic factors for patients with exocrine pancreatic cancer resectable or potentially resectable (Pancreas-CGE) | Collection of clinical, biological, and quality of life data from patients with a borderline or resectable pancreatic cancer to identify new biomarkers evaluation of treatment response and surveillance post-treatment | Prospective observational cohort | May 2023 | Recruiting |
| NCT04241367 | Verification of predictive biomarkers for pancreatic cancer treatment using multicenter liquid biopsy | Quantification and monitoring of KRAS mutations in ctDNA in pancreatic cancer pretreatment and at multiple timepoints thereafter; Discovery of biomarkers through ctDNA panel using genomic DNA and ctDNA | Prospective observational cohort | December 2025 | Recruiting |
| NCT03250078 | A pancreatic cancer screening study in hereditary high risk individuals | Screening patients at high risk for pancreatic cancer with MRI/MRCP and collection of serum to correlate biomarkers (including ctDNA) with early pancreatic cancer | Prospective observational cohort | November 2026 | Recruiting |
ctDNA may also enhance pancreatic cancer post-treatment surveillance. In the previously described prospective study, Pietrasz and colleagues collected longitudinal plasma samples at various time points (every 10-30 days) in a subgroup of 8 patients. They noted that ctDNA levels correlated with radiologic change, and progression of disease was detected at a median of 2.4 months using ctDNA compared to 4 months with CT scan surveillance[33]. Similarly, in a study containing nine patients with postoperative cancer recurrence, ctDNA levels were detectable at an average of 3.1 months, compared to 9.6 months for detectable change on CT scan (n = 9, P = 0.0004, paired t-test)[40].
CONCLUSION
Despite advances in surgical management and chemotherapy, pancreatic cancer remains a difficult disease to treat. Given its often late presentation and high rates of distant recurrence, biomarkers are needed to enhance surveillance and treatment decision making. ctDNA may be a key biomarker to better understand the burden of disease and guide neoadjuvant and adjuvant therapies in pancreatic cancer. Utilizing ctDNA assessment to identify early-stage disease, guide therapy based on risk profile, and monitor for early recurrence may also have an impact on survival. Refinement of ctDNA detection techniques to make this biomarker more scalable and affordable to test, and more experience with its interpretation during the natural history of the disease, may affirm ctDNA as a useful biomarker in pancreatic cancer.
DECLARATIONS
Authors’ contributionsMade substantial contributions to conception and design of the study and performed article analysis and interpretation: Tyerman Z, Ambler E, Chawla A
Provided administrative, technical, and editorial support: Schlick CJR, Nwajei F
Availability of data and materialsNot applicable.
Financial support and sponsorshipNone.
Conflicts of interestAll authors declared that there are no conflicts of interest.
Ethical approval and consent to participateNot applicable.
Consent for publicationNot applicable.
Copyright© The Author(s) 2022.
REFERENCES
1. Centers for Disease Control and Prevention, An update on cancer deaths in the United States, 2021. Available from: https://www.cdc.gov/cancer/dcpc/research/update-on-cancer-deaths/index.htm [Last accessed on 20 July 2022].
2. American Cancer Society, Cancer facts & figures 2021. Available from: https://www.cancer.org/research/cancer-facts-statistics/all-cancer-facts-figures/cancer-facts-figures-2021.html [Last accessed on 20 July 2022].
3. McGuigan A, Kelly P, Turkington RC, Jones C, Coleman HG, McCain RS. Pancreatic cancer: a review of clinical diagnosis, epidemiology, treatment and outcomes. World J Gastroenterol 2018;24:4846-61.
4. Moletta L, Serafini S, Valmasoni M, Pierobon ES, Ponzoni A, Sperti C. Surgery for recurrent pancreatic cancer: is it effective? Cancers (Basel) 2019;11:991.
5. Komo T, Murakami Y, Kondo N, et al. Prognostic impact of para-aortic lymph node micrometastasis in pancreatic ductal adenocarcinoma. Ann Surg Oncol 2016;23:2019-27.
6. Niedergethmann M, Rexin M, Knob S, et al. [Detection of micrometastases after curative resection for ductal adenocarcinoma of the pancreas]. Zentralbl Chir 2001;126:917-21.
7. Ferrone CR, Finkelstein DM, Thayer SP, Muzikansky A, Fernandez-delCastillo C, Warshaw AL. Perioperative CA19-9 levels can predict stage and survival in patients with resectable pancreatic adenocarcinoma. J Clin Oncol 2006;24:2897-902.
8. Pavai S, Yap SF. The clinical significance of elevated levels of serum CA 19-9. Med J Malaysia 2003;58:667-72.
9. Kim JE, Lee KT, Lee JK, Paik SW, Rhee JC, Choi KW. Clinical usefulness of carbohydrate antigen 19-9 as a screening test for pancreatic cancer in an asymptomatic population. J Gastroenterol Hepatol 2004;19:182-6.
10. Ballehaninna UK, Chamberlain RS. Serum CA 19-9 as a biomarker for pancreatic cancer-a comprehensive review. Indian J Surg Oncol 2011;2:88-100.
11. Ballehaninna UK, Chamberlain RS. The clinical utility of serum CA 19-9 in the diagnosis, prognosis and management of pancreatic adenocarcinoma: An evidence based appraisal. J Gastrointest Oncol 2012;3:105-19.
12. Duffy MJ, Sturgeon C, Lamerz R, et al. Tumor markers in pancreatic cancer: a European Group On Tumor Markers (EGTM) status report. Ann Oncol 2010;21:441-7.
13. Mandel P, Metais P. Nuclear acids in human blood plasma. C R Seances Soc Biol Fil 1948;142:241-3.
14. Leon SA, Shapiro B, Sklaroff DM, Yaros MJ. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res 1977;37:646-50.
15. Elazezy M, Joosse SA. Techniques of using circulating tumor DNA as a liquid biopsy component in cancer management. Comput Struct Biotechnol J 2018;16:370-8.
16. Gorgannezhad L, Umer M, Islam MN, Nguyen NT, Shiddiky MJA. Circulating tumor DNA and liquid biopsy: opportunities, challenges, and recent advances in detection technologies. Lab Chip 2018;18:1174-96.
17. Hindson BJ, Ness KD, Masquelier DA, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem 2011;83:8604-10.
18. Myllykangas S, Ji HP. Targeted deep resequencing of the human cancer genome using next-generation technologies. Biotechnol Genet Eng Rev 2010;27:135-58.
19. Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014;6:224ra24.
20. Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med 2008;14:985-90.
21. Konishi F. CEA doubling time and CEA half-life in the prediction of recurrences after colorectal cancer surgery. Jpn J Clin Oncol 2002;32:41-2.
22. Dawson SJ, Tsui DW, Murtaza M, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med 2013;368:1199-209.
23. Diaz LA Jr, Williams RT, Wu J, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature 2012;486:537-40.
24. Misale S, Yaeger R, Hobor S, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature 2012;486:532-6.
25. Siravegna G, Mussolin B, Buscarino M, et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat Med 2015;21:795-801.
26. Kim YW, Kim YH, Song Y, et al. Monitoring circulating tumor DNA by analyzing personalized cancer-specific rearrangements to detect recurrence in gastric cancer. Exp Mol Med 2019;51:1-10.
27. Siravegna G, Bardelli A. Blood circulating tumor DNA for non-invasive genotyping of colon cancer patients. Mol Oncol 2016;10:475-80.
28. Tie J, Kinde I, Wang Y, et al. Circulating tumor DNA as an early marker of therapeutic response in patients with metastatic colorectal cancer. Ann Oncol 2015;26:1715-22.
29. Mohan S, Ayub M, Rothwell DG, et al. Analysis of circulating cell-free DNA identifies KRAS copy number gain and mutation as a novel prognostic marker in Pancreatic cancer. Sci Rep 2019;9:11610.
30. Sugimori M, Sugimori K, Tsuchiya H, et al. Quantitative monitoring of circulating tumor DNA in patients with advanced pancreatic cancer undergoing chemotherapy. Cancer Sci 2020;111:266-78.
31. Kruger S, Heinemann V, Ross C, et al. Repeated mutKRAS ctDNA measurements represent a novel and promising tool for early response prediction and therapy monitoring in advanced pancreatic cancer. Ann Oncol 2018;29:2348-55.
32. Lee B, Lipton L, Cohen J, et al. Circulating tumor DNA as a potential marker of adjuvant chemotherapy benefit following surgery for localized pancreatic cancer. Ann Oncol 2019;30:1472-8.
33. Pietrasz D, Pécuchet N, Garlan F, et al. Plasma circulating tumor DNA in pancreatic cancer patients is a prognostic marker. Clin Cancer Res 2017;23:116-23.
34. Groot VP, Mosier S, Javed AA, et al. Circulating tumor DNA as a clinical test in resected pancreatic cancer. Clin Cancer Res 2019;25:4973-84.
35. Kim MK, Woo SM, Park B, et al. Prognostic implications of multiplex detection of. KRAS ;64:726-34.
36. Däbritz J, Preston R, Hänfler J, Oettle H. Follow-up study of K-ras mutations in the plasma of patients with pancreatic cancer: correlation with clinical features and carbohydrate antigen 19-9. Pancreas 2009;38:534-41.
37. Steinberg WM, Gelfand R, Anderson KK, et al. Comparison of the sensitivity and specificity of the CA19-9 and carcinoembryonic antigen assays in detecting cancer of the pancreas. Gastroenterology 1986;90:343-9.
38. Wong T, Howes N, Threadgold J, et al. Molecular diagnosis of early pancreatic ductal adenocarcinoma in high-risk patients. Pancreatology 2001;1:486-509.
Cite This Article
Export citation file: BibTeX | RIS
OAE Style
Tyerman Z, Ambler E, Schlick CJR, Nwajei F, Chawla A. Utility of circulating tumor DNA for predicting prognosis in the management of resectable pancreatic cancer . J Cancer Metastasis Treat 2022;8:29. http://dx.doi.org/10.20517/2394-4722.2022.20
AMA Style
Tyerman Z, Ambler E, Schlick CJR, Nwajei F, Chawla A. Utility of circulating tumor DNA for predicting prognosis in the management of resectable pancreatic cancer . Journal of Cancer Metastasis and Treatment. 2022; 8: 29. http://dx.doi.org/10.20517/2394-4722.2022.20
Chicago/Turabian Style
Tyerman, Zachary, Emily Ambler, Cary Jo R. Schlick, Felix Nwajei, Akhil Chawla. 2022. "Utility of circulating tumor DNA for predicting prognosis in the management of resectable pancreatic cancer " Journal of Cancer Metastasis and Treatment. 8: 29. http://dx.doi.org/10.20517/2394-4722.2022.20
ACS Style
Tyerman, Z.; Ambler E.; Schlick CJR.; Nwajei F.; Chawla A. Utility of circulating tumor DNA for predicting prognosis in the management of resectable pancreatic cancer . J. Cancer. Metastasis. Treat. 2022, 8, 29. http://dx.doi.org/10.20517/2394-4722.2022.20
About This Article
Special Issue
Copyright
Data & Comments
Data

 Cite This Article 8 clicks
Cite This Article 8 clicks











Comments
Comments must be written in English. Spam, offensive content, impersonation, and private information will not be permitted. If any comment is reported and identified as inappropriate content by OAE staff, the comment will be removed without notice. If you have any queries or need any help, please contact us at support@oaepublish.com.