Radiomics in sarcoma trials: a complement to RECIST for patient assessment
Abstract
Radiological imaging has a critical role in the diagnosis of sarcomas and in evaluating therapy response assessment. The current gold standard for response assessment in solid tumors is the Response Evaluation Criteria in Solid Tumors, which evaluates changes in tumor size as a surrogate endpoint for therapeutic efficacy. However, tumors may undergo necrosis, changes in vascularization or become cystic in response to therapy, with no significant volume changes; thus, size assessments alone may not be adequate. Such morphological changes may give rise to radiographic phenotypes that are not easily detected by human operators. Fortunately, recent advances in high-performance computing and machine learning algorithms have enabled deep analysis of radiological images to extract features that can provide richer information about intensity, shape, size or volume, and texture of tumor phenotypes. There is growing evidence to suggest that these image-derived or “radiomic features” are sensitive to biological processes such as necrosis and glucose metabolism. Thus, radiomics could prove to be a critical tool for assessing treatment response and may present an integral complement to existing response criteria, opening new avenues for patient assessment in sarcoma trials.
Keywords
INTRODUCTION
In the era of precision oncology, it is crucial to robustly evaluate therapeutic efficacy. In clinical trials, this evaluation is applied under predefined conditions to test new agents and quantify their level of antitumor activity. Methodology to evaluate treatment response has greatly evolved over the past decades, from a subjective evaluation reported by the treating physician to a complex set of objective criteria attempting to standardize the response evaluation process[1-3]. Traditional or gold standard response criteria are based on changes in tumor size as measured on radiological imaging. However, size criteria alone may be insufficient when assessing response in certain tumors, such as soft tissue sarcomas (STSs). In addition to being potentially difficult to measure, some STSs may not change in size in response to therapy, at least initially[1].
The World Health Organization (WHO) first established a standardized approach to evaluating treatment responses of solid tumors based on size changes[2,4]. For all intents and purposes, the WHO criteria were a precursor to the Response Evaluation Criteria in Solid Tumors (RECIST)[2,5]. Tumors were to be selected and measured in two dimensions (cross-product of the longest diameter and the longest perpendicular diameter) using a ruler or calipers; percent increases or decreases in the sum of areas would indicate progressive disease or partial response (PR), respectively[4]. However, the WHO criteria had its limitations. Notably, variance among research groups and the arrival of new imaging technologies [computed tomography (CT) and magnetic resonance imaging (MRI)] led to some confusion about how to integrate three-dimensional measures into response assessment[6]. Issues such as these necessitated clarification and modification of the WHO criteria, which ultimately led to the inception of RECIST in the year 2000.
The changes introduced by RECIST represented an improvement from WHO criteria[6]. Measuring all lesions in two dimensions was time-consuming, with a high risk of measurement error. Instead, RECIST used a one-dimensional measurement of lesion diameter. The limitations of measurable lesions were clarified in RECIST as well. Up to 10 lesions (maximum of 5 lesions/organ) could be measured, provided they met minimum size requirements (e.g., a diameter of 10 mm on spiral CT or 20 mm on non-spiral CT or MRI). With RECIST 1.1 came even more improvements, such as the inclusion of functional imaging and measurement definition of pathologic lymph nodes.
Currently, RECIST 1.1 is the gold standard for the radiological assessment of treatment response in solid tumors[6,7]. It forms the basis for progression-free survival determination and defines progressive disease as at least a 20% increase in the sum of diameters of up to five target lesions (maximum of two lesions/organ), taking as reference the smallest sum of diameters on the study and an absolute lesion increase of at least
Limitations of RECIST
While RECIST represents a common language of treatment efficacy for clinical researchers across disease sites and clinical trial settings, it has limitations. Systemic therapy with traditional chemotherapeutic agents is a cornerstone of sarcoma treatment. These agents are cytotoxic and act primarily through the inhibition of cell division. Growth inhibition of neoplastic cells is indicated by a change in tumor size hence its use as a radiographic biomarker of treatment response. However, in response to systemic therapy, a sarcoma may undergo necrosis, change in vascularization or become cystic, with no significant change in tumor size[8]. Likewise, while cytotoxic agents remain the mainstay for most sarcomas, the use of targeted therapies is increasingly common. Molecular-targeted therapy and immunotherapies may induce changes associated with an improvement in outcomes that do not correlate with an immediate change in tumor size. A notable example of this is gastrointestinal stromal tumors (GISTs), where the Choi response criteria are more sensitive and more precise than RECIST in assessing response to imatinib mesylate[9] through the inclusion of tumor density changes[10]. Irrespective of treatment, the one-size-fits-all approach of measuring diameter changes is simply not an optimal criterion for assessing response to therapy in sarcomas, given their heterogeneity in response.
Another limitation of RECIST is that, within its framework, for patients with multiple lesions, the selection of target lesions in different organs may vary between operators. Further, linear measurements of tumor size may have limitations related to variability in technical and imaging factors, tumor enhancement and morphology, and reader decisions. These variational factors pose a challenge when comparing tumor size changes throughout the course of a clinical trial. One solution would be to measure the entire tumor volume, which would subsequently improve our ability to reliably detect small changes in lesion measurements. Currently, volume-based measurement of lesions is not included in RECIST because of limitations in past diagnostic imaging techniques and available methods of measurement. However, improvements in imaging hardware and increasingly-available tumor segmentation software can turn the possibility of measuring tumor volumes into a reality.
Radiomics to the rescue
Imaging procedures produce large volumes of digital imaging data from regional to whole-body scans, capturing different aspects of human anatomy and physiology. Despite their outward complexity, however, radiological images are simply a collection of two-dimensional arrays of numeric values. Inherently a quantitative construct, these images present an opportunity for mathematical manipulation of their constituent numeric values - the data contained within the image. Radiomics is an emerging field that converts imaging data into quantitative imaging features, also called “radiomic features”[11,12]. These features may provide richer information about intensity, shape, size or volume, and texture of tumor phenotypes that may or may not be overtly perceptible to the human eye[13].
The computation of radiomic features relies on lesion segmentation - that is, the location and subsequent delineation of boundaries of the said lesion in a medical image[14]. In the context of radiation therapy, manual segmentation is routinely performed by experts to define the treatment target and normal structures[14]. Similar to the choice and measurement of target lesions in RECIST 1.1, manual delineation is time-consuming and subjective. The effect of inter- and intra-observer segmentation variability and its effect on downstream radiomic analyses has been described extensively[15]. In an effort to minimize this variability and thereby promote the reproducibility and translatability of radiomic studies, segmentation can also be performed semi-automatically (using established techniques such as region growing) or fully-automatically (using deep learning algorithms)[16]. With improvements in algorithms and computer hardware (i.e., graphical processing units or GPUs) in recent years, fully-automatic segmentation using deep learning has begun to establish itself as state-of-the-art for medical image segmentation[16]. Algorithms have been applied to auto-segment targets and normal tissues in many anatomical sites, including the thorax, abdomen, pelvis, head and neck, and brain[16]. Some applications have produced better results than the measured inter- and intraobserver contouring variability, which may lend itself to increased acceptance and adoption of fully-automated segmentation into clinical practice. This, in theory, presents the opportunity to interrogate all lesions above a certain size within selected organs, as opposed to limiting assessments to selected lesions.
Radiomics for STS
Radiomics has been deployed for the prediction of either diagnosis or prognosis in STS. Specific tasks include benign versus malignant tumor discrimination, grading, tumor histotype discrimination, survival, local and/or metastatic relapse and response to therapy. The latter, although relatively unexplored in STS, can complement existing response criteria through the interrogation of other potentially valuable metrics. Tian et al. evaluated the role of CT texture analysis in assessing the pathological response of STS treated with neoadjuvant immunotherapy plus radiotherapy (RT) in comparison to tumor size, density, and perfusion[17]. The percent change in a CT-derived textural feature after eight weeks had a significant correlation with tumor necrosis in surgical specimens, whereas those of size, density, and perfusion did not. This would suggest that textural metrics may be sensitive to underlying tumor biology. Towards a related endpoint, Escobar et al. revisited a dataset originally presented for the prediction of metastatic relapse[18,19]. The authors produced quantitative maps within the tumor to highlight the signal that contributes to decision making within a predictive framework. Analysis of these maps identified two biological patterns that are consistent with STS grading systems and knowledge: necrosis development and glucose metabolism of the tumor. Crombé et al. evaluated heterogeneity in tumor vascularization through texture analysis towards improved predictions of patients’ outcomes and response evaluation[20]. In their analysis of 25 STS patients, the area under the receiver operating characteristics curve (AUC) of predictive models ranged from 0.77 to 0.90(R2-1).
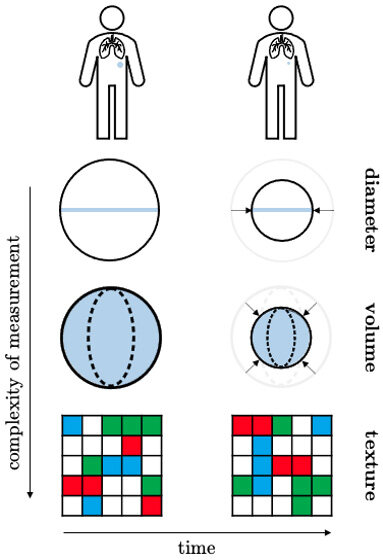
Traditional radiomics approaches use images collected at a single time point (i.e., at baseline), ignoring the changes that occurred during treatment or subsequently in follow-up[21]. The radiomic features from this single image are typically linked to clinical and biological endpoints. Nevertheless, in response to treatment, a tumor may undergo morphological changes, such as fibrotic or necrotic processes. These processes may bring about a change in tumor heterogeneity, ultimately changing the data contained within the image. Delta-radiomics quantifies the change in radiomics features (and hence change in intratumoral heterogeneity) during or after treatment, providing additional information about tumor response to treatment [Figure 1]. Recent studies have highlighted the utility of a delta-radiomics approach for differential diagnosis, survival estimation and the evaluation of treatment response[22]. A retrospective study by Crombé et al. investigated the potential of an MRI-based delta-radiomics approach to improving early response assessment in 65 high-grade STS patients treated with neoadjuvant chemotherapy. In the training cohort (n = 50), the best performance was obtained with a random forest model that was able to predict early response better than RECIST 1.1 with an AUC of 0.86, accuracy = 88.1%, sensitivity = 94.1%, and specificity = 66.3% at cross-validation. The test cohort achieved higher sensitivity and specificity but lower AUC and accuracy values (98.0%, 27.8%, 0.63% and 74.6%, respectively)[23]. Lin et al. proposed a CT-based delta-radiomics nomogram for the evaluation of pathologic response in 191 high-grade osteosarcomas treated with neoadjuvant chemotherapy with promising results[24]. Eight delta-radiomics features differed significantly between response categories with AUC 0.871 (95%CI: 0.804-0.923) in the training cohort
INTEGRATING RADIOMICS INTO CLINICAL TRIALS
By nature, clinical trials are designed to generate primary evidence on treatment efficacy, effectiveness and safety. A significant advantage of integrating radiomics analyses into clinical trials is that they leverage routinely collected imaging studies that are used to evaluate treatment efficacy. However, whether radiomic analysis is included as a trial objective (exploratory or otherwise) or considered as a contingency, it is important to be aware of the translational challenges in radiomics. Challenges include a lack of reproducibility and interpretability as well as over-fitting on small datasets, i.e., the predictive value is poor on new data. The latter, in particular, rings true in the analysis of STS patients, who are often found in smaller datasets due to the rarity of the cancer.
While the advent of publicly-available imaging datasets helps to address the sample size challenge, reproducibility and interpretability remain active areas of research in radiomics. Several initiatives and/or investigator-driven studies have proposed frameworks for analysis and quality control for enhanced reproducibility[26-31]. Arguably the greatest challenge for radiomics in sarcoma trials, however, is the generalizability of radiomic features across centers. Most imaging studies have been limited to analyses on single-center, homogeneous datasets carefully constructed using acquisition protocols that aim to minimize instrument-related variability in the data. All the same, the rarity and heterogeneity of STS pose a challenge to the design and conduct of adequately powered clinical trials[1]; hence, patients are often pooled from multiple centers for sufficient power. Variability in scanner hardware and acquisition protocols is unavoidable in this context.
This clearly presents a quantitative challenge as radiomic features are sensitive to this type of variation[15], which can be compounded with intra-center inconsistencies. One potential solution to minimize this variation would be to update the prospective trial protocol with a standardized imaging protocol. It is a commonplace for trial protocols to state when a patient should be imaged and how their disease should be monitored. As an example, a protocol may specify the imaging modality (i.e., CT or MRI), the method of assessment (i.e., RECIST 1.1), and for serial assessments, stipulate that the same imaging modality be utilized for all assessments. Of course, from a logistical standpoint, there is no guarantee that, within the same center, a single patient will be imaged using the same scanner for multiple assessments or multiple patients will be imaged using the same scanner for a single assessment. It would be a fallacy to suggest that imaging all patients on the same scanner for all assessments would be a trivial undertaking as well; however, even the incorporation of similar reconstruction techniques across centers would greatly improve the generalizability of radiomics features, which would increase their potential for clinical translatability.
New horizons
While radiomic features are typically linked to clinical and biological endpoints, there are growing research efforts to integrate radiological parameters with other information extracted from different sources. Radiogenomics is a notable example of such integration, where imaging characteristics and corresponding gene expression profiles are combined to identify optimal markers of treatment response and patient prognosis[11]. Radiology and pathology correlations have also been explored, in which diagnostic information from tissue is aligned with medical imaging[32,33].
Clinical trials are becoming increasingly complex, with large amounts of diverse data being collected. Before initiating treatment on any trial, patients are screened; this generates a plethora of information in the process, including, but not limited to demographics, performance status, medical history, plasma, stool and tissue collection, swab samples and tumor assessments with imaging. These different types of data can potentially offer alternative and complementary data streams to assess treatment response in addition to radiomics. Evaluating the tumor microbiome is a promising new approach for improving our understanding of treatment efficacy. Tinoco et al. found a specific relationship between microbial presence and histological sarcoma subtype, which also statistically correlated with overall survival[34]. Liquid biopsy and subsequent interrogation of levels of ctDNA and next-generation sequencing can also offer a non-invasive means that could be leveraged for treatment response assessment. Madanat-Harjuoja et al. demonstrated that detection of ctDNA is associated with outcome and objective response to chemotherapy in patients with advanced LMS[35]. While integrating these complementary data with radiological parameters remains an open challenge, the development of such integrative biomarkers is an exciting new frontier.
CONCLUSION
Substantial effort was required to develop the initial RECIST criterion and its subsequent modifications. While size-based criteria like RECIST are often used and historically accepted because of their relative simplicity, anatomic tumor response criteria are not without limitations. Sarcomas are a wildly diverse group of cancers that may not change in size in response to therapy. Hence, a one-size-fits-all approach to measuring diameter changes is not an optimal criterion for assessing response to therapy. Imaging-based response evaluation has a pivotal role in the evolution of treatment response evaluation. While unclear what the future will hold, it should be interesting to see how response criteria evolve in the wake of improved imaging technologies and algorithms. At the confluence of these improvements lies the ability to extract potentially critical data for treatment response evaluation. We would be remiss to ignore the potential.
DECLARATIONS
AcknowledgmentsThe authors wish to thank Larry Baker and Denise Reinke for their continued support, encouragement and patience. Specifically, the authors wish to thank Larry Baker for providing inspiration, guidance and a sounding board when required.
Authors’ contributionsDesigned the contents: Geady C, Haibe-Kains B
Drafted the initial version of the manuscript: Geady C
Participated in the writing of the manuscript: Shultz DB, Razak ARA, Schuetze S, Haibe-Kains B
Availability of data and materialsNot applicable.
Financial support and sponsorshipThis research was supported by the Sarcoma Alliance for Research through Collaboration LMSARC research fund and the philanthropic LMS360 research fund from the University of Michigan LMS360.
Conflicts of interestBenjamin Haibe-Kains is a paid consultant and shareholder of Code Ocean Inc., is part of the Scientific Advisory Board of the Break through Cancer (BTC) Foundation, and is a founding member of the Massive Analysis and Quality Control (MAQC) society. Other authors declared that they have no conflicts of interest.
Ethical approval and consent to participateNot applicable.
Consent for publicationNot applicable.
Copyright© The Author(s) 2022.
REFERENCES
1. Jaffe CC. Response assessment in clinical trials: implications for sarcoma clinical trial design. Oncologist 2008;13:14-8.
2. Ko CC, Yeh LR, Kuo YT, Chen JH. Imaging biomarkers for evaluating tumor response: RECIST and beyond. Biomark Res 2021;9:52.
5. Nishino M, Jagannathan JP, Ramaiya NH, Van den Abbeele AD. Revised RECIST guideline version 1.1: what oncologists want to know and what radiologists need to know. AJR Am J Roentgenol 2010;195:281-9.
6. Therasse P, Arbuck SG, Eisenhauer EA, et al. New guidelines to evaluate the response to treatment in solid tumors. Breast Cancer 2005;12:S16-27.
7. Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 2009;45:228-47.
8. Fanciullo C, Gitto S, Carlicchi E, Albano D, Messina C, Sconfienza LM. Radiomics of musculoskeletal sarcomas: a narrative review. J Imaging 2022;8:45.
9. Benjamin RS, Choi H, Macapinlac HA, et al. We should desist using RECIST, at least in GIST. J Clin Oncol 2007;25:1760-4.
10. Choi H, Charnsangavej C, Faria SC, et al. Correlation of computed tomography and positron emission tomography in patients with metastatic gastrointestinal stromal tumor treated at a single institution with imatinib mesylate: proposal of new computed tomography response criteria. J Clin Oncol 2007;25:1753-9.
11. Aerts HJ, Velazquez ER, Leijenaar RT, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 2014;5:4006.
12. Lambin P, Rios-Velazquez E, Leijenaar R, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 2012;48:441-6.
13. Avanzo M, Wei L, Stancanello J, et al. Machine and deep learning methods for radiomics. Med Phys 2020;47:e185-202.
14. Sharp G, Fritscher KD, Pekar V, et al. Vision 20/20: perspectives on automated image segmentation for radiotherapy. Med Phys 2014;41:050902.
15. Reiazi R, Abbas E, Famiyeh P, et al. The impact of the variation of imaging parameters on the robustness of computed tomography radiomic features: a review. Comput Biol Med 2021;133:104400.
16. Cardenas CE, Yang J, Anderson BM, Court LE, Brock KB. Advances in auto-segmentation. Semin Radiat Oncol 2019;29:185-97.
17. Tian F, Hayano K, Kambadakone AR, Sahani DV. Response assessment to neoadjuvant therapy in soft tissue sarcomas: using CT texture analysis in comparison to tumor size, density, and perfusion. Abdom Imaging 2015;40:1705-12.
18. Escobar T, Vauclin S, Orlhac F, et al. Voxel-wise supervised analysis of tumors with multimodal engineered features to highlight interpretable biological patterns. Med Phys 2022;49:3816-29.
19. Vallières M, Freeman CR, Skamene SR, El Naqa I. A radiomics model from joint FDG-PET and MRI texture features for the prediction of lung metastases in soft-tissue sarcomas of the extremities. Phys Med Biol 2015;60:5471-96.
20. Crombé A, Saut O, Guigui J, Italiano A, Buy X, Kind M. Influence of temporal parameters of DCE-MRI on the quantification of heterogeneity in tumor vascularization. J Magn Reson Imaging 2019;50:1773-88.
21. Lambin P, Leijenaar RTH, Deist TM, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 2017;14:749-62.
22. Nardone V, Reginelli A, Grassi R, et al. Delta radiomics: a systematic review. Radiol Med 2021;126:1571-83.
23. Crombé A, Périer C, Kind M, et al. T2-based MRI delta-radiomics improve response prediction in soft-tissue sarcomas treated by neoadjuvant chemotherapy. J Magn Reson Imaging 2019;50:497-510.
24. Lin P, Yang PF, Chen S, et al. A Delta-radiomics model for preoperative evaluation of Neoadjuvant chemotherapy response in high-grade osteosarcoma. Cancer Imaging 2020;20:7.
25. Gao Y, Kalbasi A, Hsu W, et al. Treatment effect prediction for sarcoma patients treated with preoperative radiotherapy using radiomics features from longitudinal diffusion-weighted MRIs. Phys Med Biol 2020;65:175006.
26. Sanduleanu S, Woodruff HC, de Jong EEC, et al. Tracking tumor biology with radiomics: a systematic review utilizing a radiomics quality score. Radiother Oncol 2018;127:349-60.
27. Zwanenburg A, Vallières M, Abdalah MA, et al. The image biomarker standardization initiative: standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology 2020;295:328-38.
28. van Griethuysen JJM, Fedorov A, Parmar C, et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res 2017;77:e104-7.
29. Shaikh FA, Kolowitz BJ, Awan O, et al. Technical challenges in the clinical application of radiomics. JCO Clin Cancer Inform 2017;1:1-8.
30. Timmeren JE, Cester D, Tanadini-Lang S, Alkadhi H, Baessler B. Radiomics in medical imaging-“how-to” guide and critical reflection. Insights Imaging 2020;11:91.
31. Ibrahim A, Primakov S, Beuque M, et al. Radiomics for precision medicine: current challenges, future prospects, and the proposal of a new framework. Methods 2021;188:20-9.
32. Tang C, Hobbs B, Amer A, et al. Development of an immune-pathology informed radiomics model for non-small cell lung cancer. Sci Rep 2018;8:1922.
33. Tunali I. Hypoxia-related radiomics predict immunotherapy response: a multi-cohort study of NSCLC. bioRxiv 2020:020859.
34. Tinoco G, Husain M, Hoyd R, et al. The sarcoma microbiome as a diagnostic and therapeutic target. JCO 2021;39:11541.
Cite This Article
Export citation file: BibTeX | RIS
OAE Style
Geady C, Shultz DB, Razak ARA, Schuetze S, Haibe-Kains B. Radiomics in sarcoma trials: a complement to RECIST for patient assessment . J Cancer Metastasis Treat 2022;8:45. http://dx.doi.org/10.20517/2394-4722.2022.57
AMA Style
Geady C, Shultz DB, Razak ARA, Schuetze S, Haibe-Kains B. Radiomics in sarcoma trials: a complement to RECIST for patient assessment . Journal of Cancer Metastasis and Treatment. 2022; 8: 45. http://dx.doi.org/10.20517/2394-4722.2022.57
Chicago/Turabian Style
Geady, Caryn, David B. Shultz, Albiruni R. Abdul Razak, Scott Schuetze, Benjamin Haibe-Kains. 2022. "Radiomics in sarcoma trials: a complement to RECIST for patient assessment " Journal of Cancer Metastasis and Treatment. 8: 45. http://dx.doi.org/10.20517/2394-4722.2022.57
ACS Style
Geady, C.; Shultz DB.; Razak ARA.; Schuetze S.; Haibe-Kains B. Radiomics in sarcoma trials: a complement to RECIST for patient assessment . J. Cancer. Metastasis. Treat. 2022, 8, 45. http://dx.doi.org/10.20517/2394-4722.2022.57
About This Article
Copyright
Data & Comments
Data

 Cite This Article 7 clicks
Cite This Article 7 clicks











Comments
Comments must be written in English. Spam, offensive content, impersonation, and private information will not be permitted. If any comment is reported and identified as inappropriate content by OAE staff, the comment will be removed without notice. If you have any queries or need any help, please contact us at support@oaepublish.com.