Membrane lipid binding molecules for the isolation of bona fide extracellular vesicle types and associated biomarkers in liquid biopsy
Abstract
Cancer exacts a heavy socioeconomic cost. Earlier detection and treatment are likely to mitigate this cost. Unfortunately, conventional tissue biopsy, the gold standard in cancer diagnosis cannot fulfill the goal of earlier detection. While liquid biopsy is a promising alternative to tissue biopsy, it has its challenges and limitations. A major challenge is the isolation of bona fide lipid membrane vesicles from biological fluids. In this review, we presented a new perspective of isolating different types of extracellular vesicles (EVs) by their affinity for membrane lipid binding ligands for liquid biopsy. EVs are lipid membrane particles naturally released by almost all cells and are found in almost all biological fluids suitable for liquid biopsy. They carry materials from the secreting cells that could affect the biology of the recipient cells and could thus inform on the state and progress of the disease. However, isolating bona fide EVs is a technical challenge as biological fluids have a complex composition and contain particles or aggregates that are physically similar to EVs. Here we review the use of membrane lipid-binding ligands to isolate different bona fide EV subtypes, and to circumvent the problem of co-isolating physically similar non-EV complexes in current EV isolation protocols. We will discuss the advantages of this technique and its potential for accelerated biomarker discovery and validation through examples of pre-clinical studies. We propose that isolating EV subtypes is a technically viable and robust strategy to overcome the current bottleneck of isolating EVs for liquid biopsy.
Keywords
Introduction
Cancer is the second leading cause of death in the world according to WHO (https://www.who.int/news-room/fact-sheets/detail/cancer). In 2018, there was an estimated 18.1 million new cancer cases and 9.6 million cancer deaths worldwide with 70% occurring in low and middle-income countries[1]. This creates a devastating socioeconomic burden. To reduce this cancer burden, WHO has recommended cancer prevention through avoidance of risk factors and implementation of existing evidence-based prevention strategies. In addition, this burden can be reduced through early detection of cancer as many cancers could be cured if diagnosed early and treated adequately.
The gold standard in cancer diagnosis is tissue biopsy. However as this is an invasive procedure, it is often done when cancer is suspected and when the cancer is usually at an advanced stage. Also, the suspected cancer may be in tissues that are hard to access such as the brain and tissue biopsy will be difficult with significant clinical risks such as bleeding or infection. As such, surrogate methods for detecting the presence of cancer such as liquid biopsy are being aggressively developed. Liquid biopsy was originally defined by the US. National Cancer Institute as “a test done on a sample of blood to look for cancer cells from a tumor that are circulating in the blood or for pieces of DNA from tumor cells that are in the blood” (https://www.cancer.gov/publications/dictionaries/cancer-terms/def/liquid-biopsy). Although tissue biopsy remains the gold standard for cancer diagnosis, liquid biopsy is gaining traction as it is less invasive and could be used to screen for cancer prior to disease manifestation when the cancer is in its early stage. Biomarkers used in liquid biopsy have also expanded beyond circulating tumor cells or DNA to include circulating exosomes and miRNAs.
This review will focus primarily on the use of exosomes or more precisely, extracellular vesicles (EVs) in liquid biopsy. As recommended by Minimal Information for Studies of EVs (MISEV2014)[2] and the recently updated and expanded MISEV2018[3], the term “Exosomes” is presently used to describe small (50-200 nm) extracellular vesicle that has an endosomal biogenesis while the term “EV” is a more generic term to describe membrane vesicles secreted by cells. Since the EV types and biogenesis of most EVs in bodily fluids are presently not known, the term EV is, therefore, more appropriate when referring to secreted membrane vesicles in liquid biopsy.
EVs as sources of biomarkers
MISEV2018 defines EVs as “particles naturally released from the cell that are delimited by a lipid bilayer and cannot replicate, i.e., do not contain a functional nucleus”[3]. They are produced by nearly all cell types from bacteria to yeast, plants and animals. EVs are known to carry a complex cargo of proteins, lipids, RNA, DNA and other metabolites. As the cargo is derived from the secreting cell, the cargo reflects the type and biology of the cells and could potentially inform on the state of health in the cells. Furthermore, as EVs are generally thought to mediate intercellular communication through the conveyance of biological materials from one cell to another, EVs could potentially propagate disease or modulate host response. Hence the cargo of EVs could also predict the progress or resolution of the disease. Together, these EV features provide a strong rationale for the use of EVs in bodily fluids such as blood, urine, saliva, tears or sweat in liquid biopsy assays. The reader is referred to several recent comprehensive reviews in this area[4-8].
Advantage of plasma EV over whole plasma for biomarker discovery and analysis
EV-based liquid biopsy is essentially a transformation of the traditional assay for biomarkers in plasma or other bodily fluids into an assay for biomarkers in EVs isolated from either plasma or other bodily fluids. This transformation is critical as using EVs from bodily fluids as the source of biomarkers as opposed to unfractionated bodily fluids could greatly enhance the signal to noise ratio in detecting a biomarker. For example, plasma, a commonly used bodily fluid in diagnosis has a wide dynamic concentration range of at least 10 orders of magnitude between the most abundant plasma protein, albumin and rare biomarker proteins[9]. In fact, > 95% of the protein mass in human plasma is dominated by a dozen proteins with albumin and the globulins constituting about 50% and 25%, respectively[10]. As such, disease-associated protein biomarkers are likely to be rare and masked by the highly abundant proteins, making plasma biomarker discovery and detection challenging. Specific methods to Isolate EVs from plasma could potentially remove the highly abundant proteins from the plasma and enhances the discovery and detection of rare disease-associated biomarkers.
Technologies for isolating EVs from bodily fluids
As extensively discussed in the MISEV2018[3], EVs could be isolated from different bodily fluids such as serum/plasma, urine, tears, milk, spinal fluids using a large variety of techniques. As each of the fluids have unique features that could interfere with EV purification and analysis, MISEV emphasized the need for these fluids to be appropriately processed prior to EV isolation and analysis. For example, blood plasma is routinely treated with anti-clotting agents such as EDTA, citrate or heparin to prevent clotting while the urine is often depleted of Tamm-Horsfall protein, a major urinary protein. However, MISEV2018 does not specifically recognise any method as the gold standard for EV isolation but noted that the choice is dependent on the end use of the EVs. Presently, the most commonly used methods for EV isolation as described in MISEV2018 are generally based on biophysical parameters such as size (e.g., filtration, size exclusion chromatography, precipitation), density (e.g., differential ultracentrifugation, sucrose gradient ultracentrifugation) and charges (e.g., electrophoresis, ion exchange chromatography). EVs are also isolated according to surface proteins (e.g., immunoaffinity chromatography). Combination of these technologies, e.g., filtration with density ultracentrifugation are also being used or developed to improve the efficiency, specificity and purity of the EV preparation.
As noted by several groups, the desired end use of the purified EVs is greatly affected by the choice of isolation techniques or combination of techniques. For example, precipitation or filtration techniques could process large volumes of fluids and while this is critical in preparing sufficient therapeutic EVs, these methods may not be as stringent as size exclusion chromatography in isolating highly enriched EV preparation[11,12]. As many of the current EV isolation techniques require specialized equipment and many procedural steps, microfluidic chips are increasingly being used in the isolation of EVs as a means to reduce the hands-on time and the number of procedural steps. Most of the microfluidic chips to isolate EVs are also based on the same fractionation parameters as the conventional EV isolation techniques namely immune-capture by antibodies against surface proteins of EVs[13], size fractionation by membranes[14,15], ciliated micropillars[16], acoustics[17] or viscoelasticity[18].
Generally, most EV isolation technologies enriched for EVs on the basis of size or densities. Hence most EV preparations, especially those from biological fluids, are also significantly enriched in similar sized non-EV complexes such as HDL and LDL lipoprotein complexes and/or protein aggregates, and two or more isolation techniques often have to be used in tandem to reduce non-EV components[19-23]. In addition, most of the current EV isolation technologies require relatively large volume of sample fluid, typically 250 µL to 1 mL or long preparation time to isolate sufficient EVs for diagnostic applications.
Hence, development of EV-based liquid biopsy is hampered by two technical challenges, isolation of bona fide lipid membrane vesicles and EV isolation from small sample volumes.
Isolating EVs through their unique defining feature, lipid membrane
The unique defining feature that differentiates EVs from other secreted cellular products is their lipid membrane. Therefore, isolating EVs via their lipid membrane would enhance the specific enrichment of EVs.
Although the lipid membrane of EVs originates from the secreting cells, the EV membrane is not representative of the outer or inner cell membranes per se but rather of specific microdomains in cellular membranes. For example, the membrane of exosomes derived from Daudi cells is enriched in lipid rafts as evidenced by the presence of GM1 gangliosides which are enriched in lipid rafts[24]. It was further reported using annexin V (AV) binding studies that phosphatidylserines are exposed on the outer surface of exosomes[25]. However, it is not clear if exosomes with exposed phosphatidylserines are exosomes or apoptotic bodies as membrane vesicles from healthy cell populations are likely to contain apoptotic bodies from apoptotic cells that are inevitably present in a healthy cell population[26]. In addition, it was also observed that in platelets, AV binding activity was restricted mainly to microvesicles, and not exosomes[27]. Finally, we observed that small EVs produced by mesenchymal stem/stromal cells (MSCs) consist of at least 3 sub-types that are distinguished by their mutually exclusive binding affinity for cholera toxin B (CTB) chain, AV and shiga toxin B (ST) chain[28]. The latter three proteins bind membrane lipids, GM1 gangliosides, phosphatidylserines and globotriaosylceramides, respectively. The extraction of small EVs by these three proteins was confirmed by scanning electron microscopy images of nanosized, spherical structures of approximately 100-200 nm. Protein analysis of the three EVs revealed an enrichment of exosome-associated proteins such Alix, Tsg 101, CD9, CD81 in the CTB-binding EVs which were much reduced in the AV-binding EVs and not detectable in the ST--binding EVs. RNA was detectable only in the ST--binding EVs[28]. It was also established through pulse-chase studies that the CTB-binding EVs have an endosomal biogenesis, i.e., exosomes while the biogenesis of the AV- and ST-binding EVs is not known[28,29]. In the pulse-chase studies, the MSCs were pulse-fed with either biotinylated transferrin or biotinylated CTB and the biotinylated proteins was then “chased” to determine if the biotinylated proteins re-appear in the culture medium as EVs.
CTB, AV and ST are multi-meric with at least five lipid-binding monomers per molecule. It was reported that these monomers bind their lipid targets in a positive co-operative manner such that the lipid-bound ligand is thermodynamically most stable when all the lipid-binding monomers are bound by the target lipids[30-32]. Therefore, the binding of an EV type by any of the three ligands implies that the targeted lipids for the ligands are exposed on the EV membrane surface and are also present in high localized concentration to enable multimeric binding[28]. This thermodynamic preference of each ligand to bind multiple lipid molecules ensures that the ligand binds lipid-rich macromolecules or complexes such as EVs.
As a consequence of this preference, the lipid-binding ligand cannot be immobilized on a solid support and must be in solution to minimize steric hindrance and facilitate ligand binding to multiple lipids (unpublished observation, SKL). This thus minimizes the versatility in designing the procedural protocol for isolation.
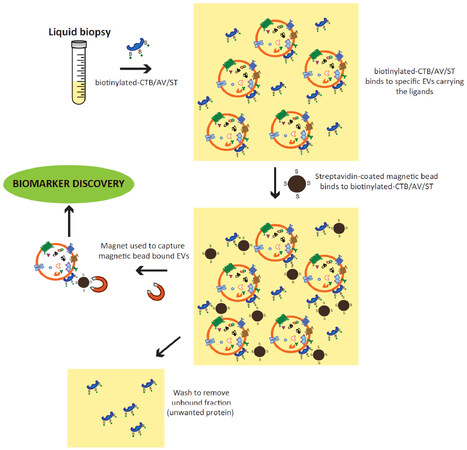
Together, the above observations suggest that EVs could be isolated through their lipid membrane via specific membrane lipid-binding ligands as shown [Figure 1].
Figure 1. Isolation of CTB/AV/ST-binding EVs from liquid biopsy. Biotinylated-CTB, AV or ST is added to the liquid biopsy. These ligands, CTB, AV or ST will bind exposed membrane lipids, GM1 gangliosides, phosphatidylserines or globotriaosylceramides on the EV membrane, respectively. After binding, the ligand-bound EVs can be readily extracted using either streptavidin-coated magnetic beads and magnets, or streptavidin-coated polystyrene and membrane fifiltration. These extracted CTB/AV/ST-binding EVs can be used directly for biomarker discovery or assay. CTB: cholera toxin B; EVs: extracellular vesicles; AV: annexin V; ST: shiga toxin
Isolating plasma EVs through their affinity for membrane lipid-binding ligands
The binding affinity of EVs for CTB and AV in conditioned medium from cell cultures has also been observed in EVs in biological fluids such as plasma and ascites fluid[33,34]. However, it was observed that less than 1% EVs in the plasma have exposed phosphatidylserines[35]. Furthermore, the CTB chain- and AV-binding EVs in the plasma and ascites fluid are distinct populations of EVs with each subtype having a cargo of proteins that is unique to EV subtype and pathological state of the patient[33,34].
Differential distribution of disease biomarkers in EV subtypes of biological fluids
When plasma EVs were isolated according to their affinity for lipid-binding ligands, it was observed that the distribution of a protein across the different EV subtypes is variable and the level of some proteins in one EV subtype but not the other EV types or the bodily fluid correlate with the disease state of the patient. This was evident even when the proteome of plasma and plasma EV sub-types from normal healthy pregnant woman and pre-eclampsia pregnant women were analyzed in low-resolution 2D protein gels[33]. When analyzed by more sensitive mass spectrometry and compared, there were differences within and between the healthy and pre-eclampsia populations of patients in each of the three fractions, namely plasma, CTB- and AV-binding plasma EVs. These differences may occur in only one or two of the fractions, and the differences could be an elevation in one fraction and a reduction in another. As such, a single source of biological fluid could yield many candidate biomarkers.
In addition, we also discovered that known proteins with weak or no correlation to a disease exhibited a strong correlation to disease when associated with a specific EV type of the biological fluid. Therefore, to facilitate the discovery of candidate biomarkers and their subsequent validation in population, we routinely screened known and well-characterized proteins for candidacy as biomarkers using antibody arrays or ELISAs[33]. The rationale for this strategy was two-fold. Firstly, the candidate biomarker would have a well characterised ELISA. Secondly, the isolation technique could be readily coupled to the standard 96 well ELISA platform leading to a semi-automated and moderately high throughput procedure[33].
As a proof of concept, we tested our strategy in a study to identify predictive biomarkers for pre-eclampsia using a prospective biobank[36]. Known and well-characterised proteins were first screened as potential pre-eclampsia biomarkers in plasma, plasma CTB-binding EV and plasma AV-binding EV from patients. Three different candidate biomarkers were identified, namely placental growth factor (PlGF), Tissue inhibitor of metalloproteinase & Plasminogen activator inhibitor-1 in plasma (plasma PlGF), plasma CTB-binding EV (CTB-TIMP1) and plasma AV-binding EV (AV-PAI1), respectively[36]. These three biomarkers were tested in a prospective biobank of 843 pregnant women. Pre-eclampsia was predicted about 7.3 (± 2.9) weeks before clinical diagnosis with a combined AUC of 0.96, sensitivity of 100%, specificity of 78.6%, and PPV of 9.9%. The cut-off concentration for predicting PE was at < 1235, ≤ 300 or > 1300 and < 10,550 pg/mL plasma for plasma PlGF, CTB-TIMP1 & AV-PAI1 respectively.
Differences in the proteome of the different EV types were also observed in cancer patients. Using a similar strategy as in the pre-eclampsia study, it was observed that MMP9 was elevated in AV-, but not detectable in CTB- or ST-binding EVs from the ascites fluid of serous ovarian cancer patients. However, this MMP9 elevation was not observed in AV-binding EVs from ascites fluid of liver cirrhosis patients[34]. More recently, a preliminary screen of plasma, plasma CTB-binding EV and plasma AV-binding EV revealed the presence of potential biomarkers that could prospectively predict the outcome of chemoradiation therapy in patients with head and neck squamous-cell carcinoma[37].
The presence of EVs in biological fluids with different affinities for membrane lipid-binding ligands demonstrated that the presence of concentrated, exposed domains of specific membrane lipids in some EV types are not unique to EVs from cell cultures. Importantly, this is a physiological phenomenon and not a consequence of the unique nutrient composition of the culture medium. It is also notable that each EV subtypes have different proteomes and that the proteome of these EV subtypes is differentially enriched in disease biomarkers.
Advantages and disadvantages of using membrane lipid binding molecules in EV isolation for diagnosis
There are several advantages of using membrane lipid binding molecules to isolate EVs for diagnosis. The most important advantage is that the principle for this isolation technique is based on the defining feature of EVs, namely the presence of a lipid membrane. Membrane lipids are generally amphiphilic, i.e., having both hydrophobic and hydrophilic ends, and in aqueous solution, they will form micelles or liposomes, i.e., vesicles. Therefore, it is likely that these membrane lipids especially when present in localized high concentrations in aqueous biological fluids, are likely to be minimally in a micelle configuration, and most possibly, a vesicular structure. The presence of membrane or cytosolic proteins will strongly corroborate the presence of membrane vesicles.
Based on affinity for membrane lipid-binding ligands, it is obvious that biological fluids contain several EV types with EV type-specific proteome. Importantly from the diagnostic perspective, this EV type-specific proteome is also dependent on the pathophysiological state of the donor and this pathophysiology-associated change in EV proteome varies with the EV type. Practically, it transforms a biological fluid as a single source of biomarkers into multiple independent sources of biomarkers.
A third advantage is that as the different EV types are novel sources of biomarkers, the distribution of known and well-characterized biomarkers and proteins in these EVs are not known. As we have shown, examining their distribution in the different EV types as a function of the pathophysiological state is a viable strategy to identify candidate biomarkers. Furthermore, it is easier to validate these known and characterized biomarkers and proteins as the necessary assay reagents such as antibodies are available.
A fourth advantage is that the technique for EV isolation by lipid-binding ligands is akin to the commonly used antibody capture techniques, and can be easily adapted for direct coupling to an ELISA for biomarker assay. Hence isolating EV from biological fluids and assaying for biomarkers by ELISA can be readily adapted for the standard automated 96 well ELISA platform used in routine clinical laboratory settings. It is also amenable to the recent developments in nano-plasmonic technology[38].
Finally, the volume of plasma required for extracting EVs by their affinity for membrane lipid binding ligands in sufficient quantity for a typical ELISA ranges from 10-30 µL biological fluids per assay[34,36]. As each of the EV types identified so far are unique EVs[28,33], the volume of biological fluid could be reduced further by using multiplexed EV isolation system where the different membrane lipid-binding ligands are added to the same sample and the bound EVs are then extracted according to the ligands bound to the EVs.
Despite the advantages of lipid binding ligands for isolation of EVs, there are significant limitations with this approach. The avidity of the ligands for the different lipids has turned out to be a double-edged sword. The dissociation constants (KD) of CTB to GM1 is between 4.6 × 10-12[39] and 7.3 × 10-10[40], KD of AV to phosphatidylserine is 5 × 10-10 M[41,42] and KD of ST to GB3 is 5-30 × 10-10 M[32]. These dissociation constants which are comparable to the median KD of antibody to antigen is 5-30 × 10-11 M[43] illustrate the difficulty in dissociating the EVs from the ligands for subsequent characterisation. While this poses significant challenge for biological studies of the different EV types, biomarker discovery and assay can be readily performed on the ligand-bound EVs.
Another drawback of this approach is the limited number of suitable lipid binding ligands to isolate EVs. Also, it is highly conceivable that many other EV types may not exhibit sufficiently distinctive lipid molecules for EV stratification by this method. Also, the lipid binding characteristics of an EV is not likely to be a characteristic of a specific cell type and is thus not likely to provide information on the cell source of the EVs. Nevertheless, lipid binding ligands represent an additional tool to isolate and stratify EV types that could increase the yield and specificity of EV types when complemented with present EV isolation tools such as size exclusion fractionation or immunoaffinity isolation technology.
Conclusion
We propose that the use of membrane lipid binding ligands in EV isolation for liquid biopsy is a viable and robust strategy to circumvent the present technical limitations in isolating bona fide EVs from biological fluids for liquid biopsy. In addition, this isolation technique is versatile can be easily integrated into the current clinical analysis platforms using 96 well assay plates and even newer analytical platforms such as nano-plasmonics[44] which uses the interaction of light and electronic oscillations in metallic substrates in a nanometer-dimension to detect the presence of single EV.
Declarations
Authors’ contributionsMade substantial contributions to conception: Lai RC, Lim SK
Provided initial drafts of the work: Lai RC
Revising the work critically for important intellectual content: Tan KH, Lim SK
Final approval of the version: Lim SK
Availability of data and materialsNot applicable.
Financial support and sponsorshipNone.
Conflicts of interestLim SK holds founder shares in Paracrine Therapeutics Pte Ltd.
Ethical approval and consent to participateNot applicable.
Consent for publicationNot applicable.
Copyright© The Author(s) 2019.
REFERENCES
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424.
2. Lötvall J, Hill AF, Hochberg F, Buzás EI, Di Vizio D, et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. J Extracell Vesicles 2014;3:26913.
3. Thery C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles 2018;7:1535750.
4. Lane RE, Korbie D, Hill MM, Trau M. Extracellular vesicles as circulating cancer biomarkers: opportunities and challenges. Clin Transl Med 2018;7:14.
5. Zhang P, Samuel G, Crow J, Godwin A, Zeng Y. Molecular Assessment of Circulating Exosomes towards Liquid Biopsy Diagnosis of Ewing Sarcoma Family of Tumors. Transl Res 2018;201:136-153.
6. Schwich E, Rebmann V. The Inner and Outer Qualities of Extracellular Vesicles for Translational Purposes in Breast Cancer. Front Immunol 2018;9:584.
7. Fujita Y, Kuwano K, Ochiya T, Takeshita F. The Impact of Extracellular Vesicle-Encapsulated Circulating MicroRNAs in lung cancer research. Biomed Res Int 2014;2014:486413.
8. Verma M, Lam TK, Hebert E, Divi RL. Extracellular vesicles: potential applications in cancer diagnosis, prognosis, and epidemiology. BMC Clin Pathol 2015;15:6.
9. Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics 2002;1:845-67.
10. Hortin GL, Sviridov D, Anderson NL. High-Abundance Polypeptides of the Human Plasma Proteome Comprising the Top 4 Logs of Polypeptide Abundance. Clin Chem 2008;54:1608-16.
11. Lobb RJ, Becker M, Wen Wen S, Wong CSF, Wiegmans AP, et al. Optimized exosome isolation protocol for cell culture supernatant and human plasma. J Extracell Vesicles 2015;4:27031.
12. Ludwig AK, De Miroschedji K, Doeppner TR, Börger V, Ruesing J, et al. Precipitation with polyethylene glycol followed by washing and pelleting by ultracentrifugation enriches extracellular vesicles from tissue culture supernatants in small and large scales. J Extracell Vesicles 2018;7:1528109.
13. Chen C, Skog J, Hsu CH, Lessard RT, Balaj L, et al. Microfluidic isolation and transcriptome analysis of serum microvesicles. Lab Chip 2010;10:505-11.
14. Davies RT, Kim J, Jang SC, Choi EJ, Gho YS, et al. Microfluidic filtration system to isolate extracellular vesicles from blood. Lab Chip 2012;12:5202-10.
15. Liang LG, Kong MQ, Zhou S, Sheng YF, Wang P, et al. An integrated double-filtration microfluidic device for isolation, enrichment and quantification of urinary extracellular vesicles for detection of bladder cancer. Sci Rep 2017;7:46224.
16. Wang Z, Wu HJ, Fine D, Schmulen J, Hu Y, et al. Ciliated micropillars for the microfluidic-based isolation of nanoscale lipid vesicles. Lab Chip 2013;13:2879-82.
17. Lee K, Shao H, Weissleder R, Lee H. Acoustic purification of extracellular microvesicles. ACS Nano 2015;9:2321-7.
18. Liu C, Guo J, Tian F, Yang N, Yan F, et al. Field-Free Isolation of Exosomes from Extracellular Vesicles by Microfluidic Viscoelastic Flows. ACS Nano 2017;11:6968-76.
19. Yuana Y, Levels J, Grootemaat A, Sturk A, Nieuwland R. Co-isolation of extracellular vesicles and high-density lipoproteins using density gradient ultracentrifugation. J Extracell Vesicles 2014;3:23262.
20. van Eijndhoven MAJ, Zijlstra JM, Groenewegen NJ, Drees EEE, van Niele S, et al. Plasma vesicle miRNAs for therapy response monitoring in Hodgkin lymphoma patients. JCI Insight 2017;1:e89631.
21. Karimi N, Cvjetkovic A, Jang SC, Crescitelli R, Hosseinpour Feizi MA, et al. Detailed analysis of the plasma extracellular vesicle proteome after separation from lipoproteins. Cell Mol Life Sci 2018;75:2873-86.
22. Sódar BW, Kittel Á, Pálóczi K, Vukman KV, Osteikoetxea X, et al. Low-density lipoprotein mimics blood plasma-derived exosomes and microvesicles during isolation and detection. Sci Rep 2016;6:24316.
23. Simonsen Jens B. What Are We Looking At? Extracellular Vesicles, Lipoproteins, or Both? Circ Res 2017;121:920-2.
24. de Gassart A, Géminard C, Février B, Raposo G, Vidal M. Lipid raft-associated protein sorting in exosomes. Blood 2003;102:4336-44.
25. Morelli AE, Larregina AT, Shufesky WJ, Sullivan ML, Stolz DB, et al. Endocytosis, intracellular sorting, and processing of exosomes by dendritic cells. Blood 2004;104:3257-66.
26. Théry C, Boussac M, Véron P, Ricciardi-Castagnoli P, Raposo G, et al. Proteomic Analysis of Dendritic Cell-Derived Exosomes: A Secreted Subcellular Compartment Distinct from Apoptotic Vesicles. J Immunol 2001;166:7309-18.
27. Heijnen HFG, Schiel AE, Fijnheer R, Geuze HJ, Sixma JJ. Activated Platelets Release Two Types of Membrane Vesicles: Microvesicles by Surface Shedding and Exosomes Derived From Exocytosis of Multivesicular Bodies and -Granules. Blood 1999;94:3791-9.
28. Lai RC, Tan SS, Yeo RW, Choo AB, Reiner AT, et al. MSC secretes at least 3 EV types each with a unique permutation of membrane lipid, protein and RNA. J Extracell Vesicles 2016;5:29828.
29. Tan SS, Yin Y, Lee T, Lai RC, Yeo RWY, et al. Therapeutic MSC exosomes are derived from lipid raft microdomains in the plasma membrane AU - Tan, Soon Sim. J Extracell Vesicles 2013;2:22614.
30. Backlund PS, Aksamit RR, Unson CG, Goldsmith P, Spiegel AM, et al. Immunochemical and electrophoretic characterization of the major pertussis toxin substrate of the RAW264 macrophage cell line. Biochemistry 1988;27:2040-6.
31. Janko C, Jeremic I, Biermann M, Chaurio R, Schorn C, et al. Cooperative binding of Annexin A5 to phosphatidylserine on apoptotic cell membranes. Phys Biol 2013;10:065006.
32. G Peter M, Lingwood C. Apparent cooperativity in multivalent verotoxin-globotriaosyl ceramide binding: Kinetic and saturation binding studies with [125I]verotoxin. Biochim Biophys Acta 2000;1501:116-24.
33. Tan KH, Tan SS, Sze SK, Lee WKR, Ng MJ, et al. Plasma biomarker discovery in preeclampsia using a novel differential isolation technology for circulating extracellular vesicles. Am J Obstet Gynecol 2014;211:380.e1-3.
34. Reiner AT, Tan S, Agreiter C, Auer K, Bachmayr-Heyda A, et al. EV-Associated MMP9 in High-Grade Serous Ovarian Cancer Is Preferentially Localized to Annexin V-Binding EVs. Dis Markers 2017;2017:9653194.
35. Arraud N, Linares R, Tan S, Gounou C, Pasquet JM, et al. Extracellular vesicles from blood plasma: determination of their morphology, size, phenotype and concentration. J Thromb Haemost 2014;12:614-27.
36. Tan KH, Tan SS, Ng MJ, Tey WS, Sim WK, et al. Extracellular vesicles yield predictive pre-eclampsia biomarkers. J Extracell Vesicles 2017;6:1408390.
37. Rodrigues-Junior DM, Tan SS, de Souza Viana L, Carvalho AL, Lim SK, et al. A preliminary investigation of circulating extracellular vesicles and biomarker discovery associated with treatment response in head and neck squamous cell carcinoma. BMC Cancer 2019;19:373.
38. Reiner AT, Ferrer NG, Venugopalan P, Lai RC, Lim SK, et al. Magnetic nanoparticle-enhanced surface plasmon resonance biosensor for extracellular vesicle analysis. Analyst 2017;142:3913-21.
39. Kuziemko GM, Stroh M, Stevens RC. Cholera Toxin Binding Affinity and Specificity for Gangliosides Determined by Surface Plasmon Resonance. Biochemistry 1996;35:6375-84.
40. MacKenzie CR, Hirama T, Lee KK, Altman E, Young NM. Quantitative Analysis of Bacterial Toxin Affinity and Specificity for Glycolipid Receptors by Surface Plasmon Resonance. J Biol Chem 1997;272:5533-8.
41. Tait JF, Gibson D, Fujikawa K. Phospholipid binding properties of human placental anticoagulant protein-I, a member of the lipocortin family. J Biol Chem 1989;264:7944-9.
42. Andree H, Reutelingsperger C, Hauptmann R, Hemker HC, Hermens WT, et al. Binding of vascular anticoagulant alpha (VAC alpha) to planar phospholipid bilayers. J Biol Chem 1990;265:4923-8.
43. Landry JP, Ke Y, Yu GL, Zhu XD. Measuring affinity constants of 1450 monoclonal antibodies to peptide targets with a microarray-based label-free assay platform. J Immunol Methods 2015;417:86-96.
Cite This Article
Export citation file: BibTeX | RIS
OAE Style
Lai RC, Tan KH, Lim SK. Membrane lipid binding molecules for the isolation of bona fide extracellular vesicle types and associated biomarkers in liquid biopsy. J Cancer Metastasis Treat 2019;5:65. http://dx.doi.org/10.20517/2394-4722.2019.011
AMA Style
Lai RC, Tan KH, Lim SK. Membrane lipid binding molecules for the isolation of bona fide extracellular vesicle types and associated biomarkers in liquid biopsy. Journal of Cancer Metastasis and Treatment. 2019; 5: 65. http://dx.doi.org/10.20517/2394-4722.2019.011
Chicago/Turabian Style
Lai, Ruenn Chai, Kok Hian Tan, Sai Kiang Lim. 2019. "Membrane lipid binding molecules for the isolation of bona fide extracellular vesicle types and associated biomarkers in liquid biopsy" Journal of Cancer Metastasis and Treatment. 5: 65. http://dx.doi.org/10.20517/2394-4722.2019.011
ACS Style
Lai, RC.; Tan KH.; Lim SK. Membrane lipid binding molecules for the isolation of bona fide extracellular vesicle types and associated biomarkers in liquid biopsy. J. Cancer. Metastasis. Treat. 2019, 5, 65. http://dx.doi.org/10.20517/2394-4722.2019.011
About This Article
Special Issue
Copyright
Data & Comments
Data

 Cite This Article 11 clicks
Cite This Article 11 clicks











Comments
Comments must be written in English. Spam, offensive content, impersonation, and private information will not be permitted. If any comment is reported and identified as inappropriate content by OAE staff, the comment will be removed without notice. If you have any queries or need any help, please contact us at support@oaepublish.com.