Epigenetic control of autophagy in women's tumors: role of non-coding RNAs
Abstract
Cancer remains the second leading cause of death worldwide and a major public health and economic issue. To reduce the burden, new approaches are necessary to diagnose the disease at early stages and improve clinical outcomes of cancer patients, for which understanding the molecular mechanisms of carcinogenesis is crucial. Autophagy is a pro-survival pathway that ensures the removal and renewal of cellular macromolecular structures, thus playing a crucial role in the maintenance of cellular homeostasis. Dysregulation of autophagy can favor chemoresistance and survival of dormant cancer cells, thus favoring cancer progression and relapse. Several studies report dysregulated expression of long non-coding RNAs and micro-RNAs acting as tumor suppressors or tumor promoters by targeting genes involved in the autophagy pathway. Here, we focus on the role played by non-coding RNAs-mediated regulation of autophagy in development and progression of cancers in women. Understanding how epigenetics can impact autophagy might open novel therapeutic strategies in the fight against cancers in women.
Keywords
Introduction
One of the hallmarks of cancer cells is represented by the enhanced signaling that lead to uncontrolled cellular growth and proliferation. To support its neoplastic growth and its sustenance under harsh and stressful conditions, cancer cells reprogram their metabolism alongside with the dynamic crosstalk with stromal cells in the tumor microenvironment[1-3].
Cellular homeostasis largely relies on autophagy, a lysosome-mediated catabolic process that accomplishes the macromolecular turnover. Altered regulation of this process may lead to the accumulation of damaged or redundant organelles, unfolded proteins, reactive oxygen species, and oncogenic molecules that favor carcinogenesis[4]. Thus, not surprisingly, autophagy is dysregulated in several cancers, among which include cancers affecting women[5-10].
Tumorigenesis is linked to genome instability, as well as to epigenetic and metabolic alterations. While normal cells show a very low rate of mutations, in malignant cells disruption of the DNA repair system compromises the surveillance machinery, leading to accumulation of spontaneous or carcinogen-induced mutations[11]. Particularly, mutations in oncogenes and in tumor suppressor genes, which control cell behavior and cell fate, allow cancer cells to escape from cell proliferation and cell motility control, avoid apoptosis, and survive under harsh metabolic conditions[1]. Besides mutations in the gene sequence that could affect the activity, the subcellular localization and the function of oncogenic or tumor suppressive proteins, and the heritable alterations of the mechanisms controlling the expression and translation (epigenetics) of oncogenes and of tumor suppressor genes also play a role in tumorigenesis. Such epigenetic mechanisms include hypermethylation/demethylation of the promoter, histone modifications (i.e., acetylation/de-acetylation), and post-transcriptional regulation by non-coding RNAs (ncRNAs)[12]. Metabolic factors (e.g., nutrient and oxygen availability), cellular stressors (e.g., oxidative stress), cell-to-cell communications, and soluble mediators (e.g., growth factors and cytokines), present in the tumor microenvironment, greatly affect the dynamic of epigenetic changes in cancer cells, resulting in their adaptation to a pro-tumorigenic environment[9,13].
ncRNAs, transcripts without encoding potential, play a pivotal epigenetic role and have been implicated in tumorigenesis by acting as tumor suppressors or tumor promoters, at transcriptional and post-transcriptional levels[14].
In this article, we review how ncRNAs may impinge on the development and progression of women’s cancers through the epigenetic control of autophagy.
Tumors affecting women at a glance
We will focus on the most frequent malignancies affecting women, including breast, ovarian, endometrial, and cervical cancers[15].
Breast cancer
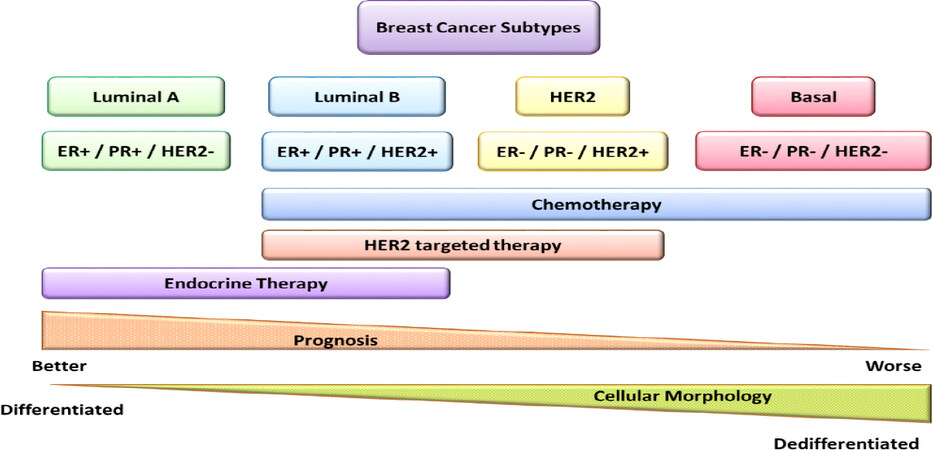
Breast cancer is the first diagnosed, in terms of incidence, and the first leading cause of death among malignancies affecting women[16]. Based on the presence or absence of predictive factors, such as estrogen or progesterone hormone receptors (HR) and human epidermal growth factor receptor 2 (HER2), breast cancer is classified into four primary subtypes: luminal A (HR+/HER2-), luminal B (HR+/HER2+), HER2 positive (HR-/HER2+), and triple-negative (HR-/HER2-). Each subtype differs in incidence, therapeutic responsiveness, and disease progression [Figure 1]. About 70% of breast cancer patients bear a luminal A or B subtype, whereas approximately 15% bear a triple-negative subtype lacking both HR and HER2 expression. HER2 is amplified in around 20% of breast cancers, and it is associated with poor prognosis[17]. HR+ tumors can be treated with tamoxifen, letrozole, anastrozole, or exemestane, while HER2+ tumors can be treated with monoclonal antibodies such as trastuzumab or pertuzumab, or with the tyrosine kinase inhibitor lapatinib. The triple negative subtype is the most aggressive among breast cancers[17], with the highest risk of relapse within 5 years[18]. Other genes frequently mutated that have a pivotal role in development and progression of breast cancer are BRCA1/2 and TP53 tumor suppressor genes[17,19,20]. The clinical outcome depends on the tumor stage at diagnosis, the presence of mutations in specific genes (e.g., BRCA1/2, TP53, PTEN) and the specific subtype, which affects the hormone-, chemo-, or molecular therapy response [Figure 1][17,21].
Ovarian cancer
Ovarian cancer is the fourth cancer, in terms of incidence, and the fourth cancer for mortality, among female malignancies[16].
The asymptomatic growth of ovarian cancer leads to a late stage diagnosis when it has already invaded the peritoneum and metastasized to distant organs[22].
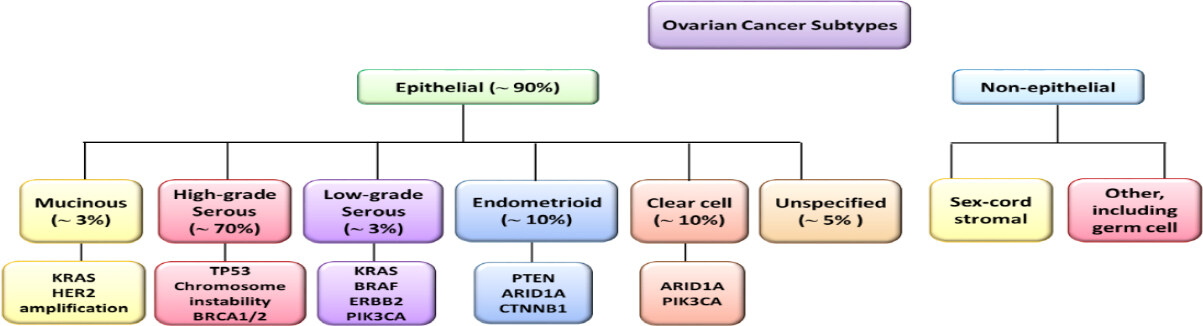
Ovarian cancers are classified as epithelial ovarian cancer (EOC), accounting for up to 90% of the total, and non-epithelial ovarian cancers, which arise from stromal and germ cells[23]. Histologically, EOCs are classified into five different subtypes: high-grade serous (HGS, accounting for 70% of the total ovarian cancers), low-grade serous (LGS, about 5%), endometrioid (about 10%), clear cell (about 10%), and mucinous (about 3%) [Figure 2][24]. In addition, some ovarian cancers present a non-specific histological pattern.
Figure 2. Ovarian cancer. Schematic representation of ovarian cancer histological types and associated gene alterations. HER2: human epidermal growth factor receptor 2; PTEN: phosphatase and tensin homolog
All these histological types, except HGS, derive from precursor lesions and grow in a stepwise manner; they are indolent and show a stable genome characterized by point mutations or amplifications and deletions in oncogenes and tumor suppressors. Serous and mucinous tumors are characterized by mutated oncogenes, such as BRAF and KRAS, whereas endometroid tumors carry PTEN mutations. Particularly, HGS ovarian cancer presents TP53 mutations in 95% of patients and germline mutations in BRCA genes in 65-85% of cases; this histological subtype is characterized by rapid growth and high aggressiveness[23,25]. Based on the genetic lesion pattern and, consequently, on the clinical features, ovarian cancers are categorized as type I or type II, the former arising from the ovary and the latter arising from the Fallopian tube or the ovarian surface epithelium[26]. Type I tumors, which include LGS, mucinous, endometrioid, clear cell, and transitional cell carcinomas, carry few mutations, are TP53 wild-type and show an indolent clinical behavior. In contrast, type II tumors, which include HGS ovarian cancer, undifferentiated carcinomas and carcinosarcomas, are genetically highly unstable with mutated TP53 and dysfunctional BRCA1/2, in the majority of cases, and show a very aggressive and metastatic behavior. Standard management of ovarian cancer comprises of a maximal cytoreductive surgery as a first line of intervention; the second therapeutic approach involves treatments with a combination of chemotherapeutic agents, such as platinum-containing drugs (cisplatin and carboplatin) and taxane drugs family (paclitaxel and docetaxel) and, as a second line, with gemcitabine, doxorubicin, and bevacizumab[27].
Endometrial cancer
Endometrial cancer is the third malignancy, in terms of incidence among female cancers[16], and the most prevalent female cancer among the American women[28].
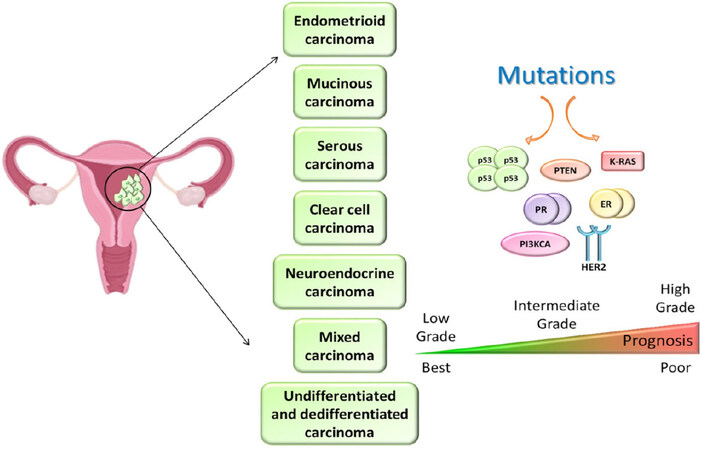
Endometrial cancers are associated with sporadic mutations of several oncogenes and tumor suppressor genes, among which PTEN, PI3KCA, KRAS, TP53, and CTNNB1 are the most frequent[29]. Women affected by the Lynch syndrome with MLH1 and MSH2 gene mutations are more susceptible to developing endometrial cancer[30]. Endometrial cancers are classified into seven different histological subtypes: (1) endometrioid carcinoma, accounting for 80% of total endometrial cancer; (2) mucinous adenocarcinoma (1%-9%); (3) serous carcinoma (less than 10%); (4) clear cell carcinoma (less than 5%); (5) neuroendocrine carcinoma; (6) mixed carcinoma; and (7) undifferentiated and dedifferentiated carcinoma[31][Figure 3]. PTEN, PIK3CA, and ER/PR are the most frequently mutated in endometrioid adenocarcinoma, while TP53 and E-cadherin are the more common mutated genes in serous carcinoma[32]. As for the management and prognosis, the majority of endometrial cancers (particularly, grades I and II endometrioid carcinomas) usually remain confined into the uterine corpus and can be removed surgically, thus having overall a good prognosis. On the contrary, serous, clear cells, grade III endometrioid, and undifferentiated carcinomas are more aggressive, often diagnosed at advanced stages with metastasis, require a more complex treatment combining surgery, radiotherapy and hormone/chemo/molecular therapies[33][Figure 3].
Cervical cancer
Cervical cancer is the second classified tumor for incidence and mortality among female malignancies[16].
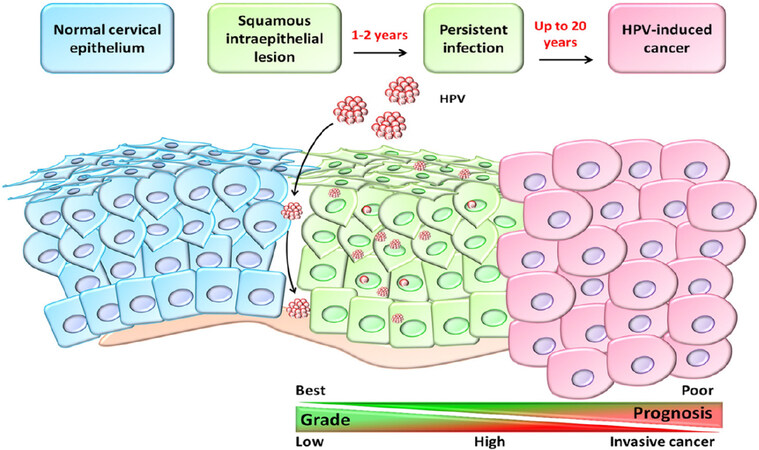
Cervical intraepithelial cells determine three grades of dysplasia, defined as cervical intraepithelial neoplasia 1 (low-grade), 2 (moderate grade), and 3 (high-grade), before becoming invasive cancer[34]. The human papilloma virus (HPV) infection is sexually transmitted and appears involved in the progression of this tumor [Figure 4]. However, the majority of HPV types are low-risk and spontaneously disappear within two years from infection. In fact, the prevalence of HPV-infection is high in women of under 25 years of age, whereas the peak of cervical cancer death occurs in middle-aged women[35]. HPV 16 and 18 are the most common oncogenic genotypes found in 75% of invasive cervical carcinoma[36]. The tumor progression is influenced by several carcinogenic factors, such as smoking, herpes simplex, HIV, and other genital virus infections[37]. In early stage, this disease is asymptomatic; nevertheless, it is strongly recommended to test for HPV in conjunction with Pap smear cytology at 21 years of age[38]. Cervical cancer treatment plan is based on clinical stage, histopathological type of tumor, grading of disease, presence of metastasis, size of primary lesion, age, and overall health status of patient. Therapeutic approaches include radical hysterectomy, radiotherapy, and/or platinum-based chemotherapy, in combination or independently[34].
Autophagy
The autophagy process at a glance
The term “autophagy” first described by Christian De Duve in 1963, derives from the Greek word for “self-eating”. Three types of autophagy have been described: macro-autophagy, micro-autophagy, and chaperone-mediated autophagy. Macro-autophagy involves the sequestration of the cargo (i.e., macromolecules and organelles that are redundant, aged, or damaged) within double membrane vacuoles, called autophagosomes, which fuse with lysosomes resulting in the breakdown of encapsulated material into simple recyclable components[39]. In the instance of micro-autophagy, the cytosolic cellular components are internalized within the lysosomes by invagination of the lysosomal membrane[40]. Chaperon-mediated autophagy is a selective and complex pathway that includes the recognition of targeted protein, containing the KFERQ motif, by the chaperone protein Hsp70, which binds to lysosomal membrane receptor lysosomal-associated membrane protein 2A (LAMP-2A) and drives the translocation of the targeted protein into the lysosome for degradation[40].
Here, we will focus on the molecular aspects of macro-autophagy, from now on simply referred to as “autophagy”. Autophagy is a catabolic process devoted to the degradation of non-functional cellular components, such as organelles (e.g., mitochondria, peroxisomes, and endoplasmic reticulum) and macromolecules (including proteins, carbohydrates, lipids, and nucleotides), within the lysosomes[41]. Extracellular and intracellular stresses, such as ER stress, pathogen infection, nutrient deprivation, hypoxia, oxidative stress, mitochondrial damage, and unfolded proteins, stimulate autophagy[42].
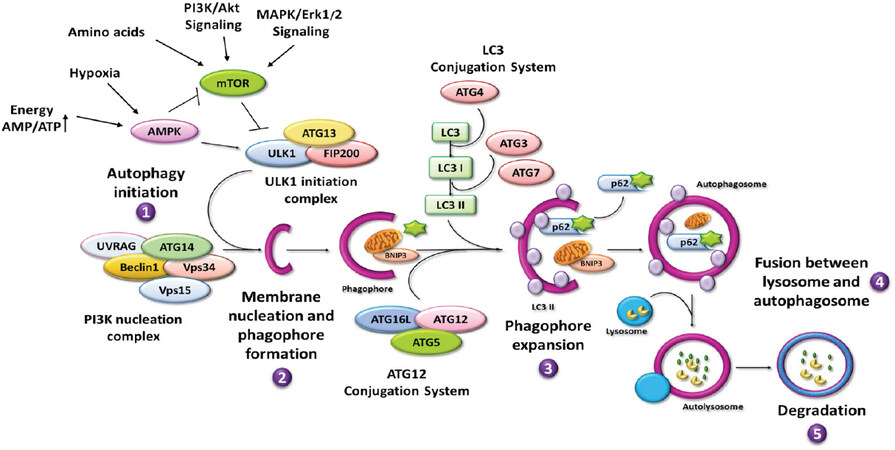
The autophagy process begins with the recognition and sequestration of the cargo within the forming autophagosome, proceeds with the autophagosome-lysosome fusion and ends with the full degradation of the cargo and translocation in the cytosol of the elementary substrates reutilized for new synthesis[43]. Ohsumi and colleagues first identified many of the autophagy-related genes (ATG) that coordinate various steps of the autophagy process, starting from the induction and subsequent formation of the autophagosome up to its fusion with the lysosome to form the autolysosome[44]. Altogether, 34 ATGs have been identified as part of the core autophagic machinery[39]. Genetic alterations involving these genes lead to autophagy dysregulation and altered cellular homeostasis, which underlie various diseases, including cancer[45]. The main steps and actors involved in the autophagy process are illustrated in Figure 5. Readers may refer to comprehensive review articles for a detailed description of the process[46-48].
Figure 5. Autophagy process. Cartoon showing a schematic overview of the main steps and autophagy-related proteins and regulators: (1) autophagy initiation; (2) membrane nucleation; (3) phagophore formation and expansion; and (4) autophagosome closure, fusion with lysosomes, and (5) degradation of cargo. ↑: increase; →: activation or induction; ⟂: inhibition; UVRAG: UV resistance-associated gene; ATG: autophagy-related gene
Briefly, the process is regulated by three kinase complexes, namely mTORC1, ULKC1, and BECLIN 1-PI3KC3 autophagy interactome[46,49]. When active, MTORC1 detaches from the lysosome and negatively acts on ULKC1, which ultimately activates the BECLIN 1-PI3KC3 complex. Thus, in the presence of growth signals that trigger the PI3KC1-AKT or the ERK/MAPK pathway as well as in the instance of abundant amino acid, mTORC1 is activated and inhibits autophagy[49,50]. On the contrary, the lack of amino acid maintains the mTORC1 attached to the lysosome membrane in the inactive state, while promoting the TFEB-mediated transcription of ATG genes[40,50]. Similarly, the lack of energetic sources and of oxygen leading to increased AMP/ATP ratio triggers AMPK that in turn inhibits mTORC1 and activates the ULKC1 and the BECLIN 1-PIK3C3 autophagy interactome[46,49]. Once active, PI3KC3 (better known as Vps34) produces PtdIns3-phosphate (PI3P), which is essential for the recruitment of membranes needed for the phagophore expansion[51]. While forming the autophagosome, the LC3-II isoform is post-translationally inserted into the bilayer of both the inner and outer membranes. Several ATG proteins (including ATGs 2, 3, 4, 5, 7, and 12) participate in processing the cytosolic isoform LC3-I to enable the conjugation with phosphatidylethanolamine, thus forming the LC3-II isoform, and for its subsequent insertion into the membranes[52]. Importantly, during the initial step of phagophore expansion, the cargo is sequestered to ensure its packaging within the autophagosome. Among the many proteins that intervenes for the cargo recognition and sequestration within the autophagosome are p62/sequestosome (p62/SQSTM1), which has a domain for binding the ubiquitinated substrate and a domain for LC3, and BNIP3, which is involved in mitochondria sequestration[46].
Epigenetics
Mechanisms at a glance
Epigenetics encompasses the biological processes involved in the regulation of gene expression without altering the nucleotide sequence; in particular, it leads to changes in phenotype without editing the genotype[53]. These mechanisms, transmissible and reversible, are pivotal to preserve the cellular homeostasis, and their alteration could favor cancer development by promoting or inhibiting oncogenic and tumor-suppressor signaling, respectively[14].
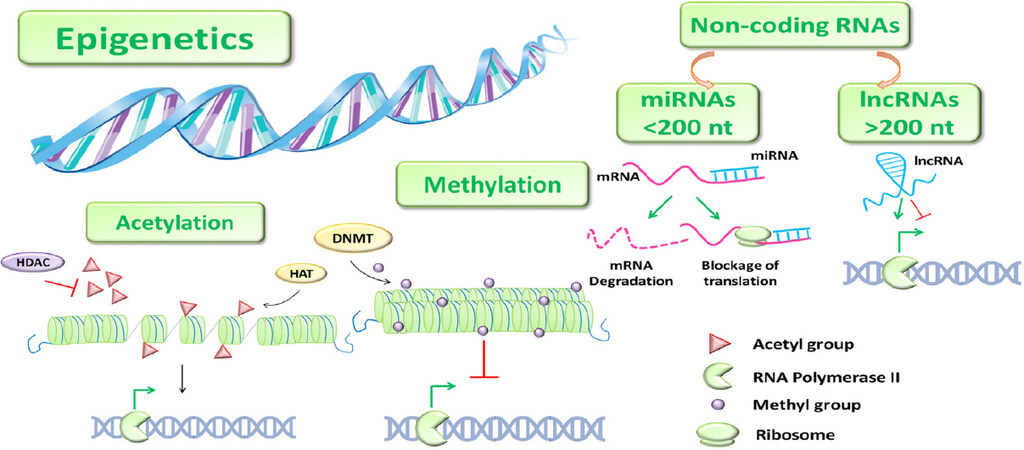
Epigenetics mainly includes chromatin remodeling that allows the transition from heterochromatin to euchromatin and vice versa, such as DNA methylation and histone modifications, and ncRNAs [Figure 6]. DNA methylation occurs in CpGs-rich regions in the promoter, leading to chromatin condensation and thus to gene transcriptional repression[54].
Figure 6. Epigenetics. Schematic DNA methyltransferase (DNMT) cartoon representing the main epigenetic mechanisms. →: activation; ⟂: inhibition; miRNAs: micro-RNAs; lncRNAs; long non-coding RNAs; HDAC: histone deacetylases
Histone modifications regard methylation, which positively and negatively interferes on gene activation, and acetylation, that favors the transcriptional activation. The latter mechanism is mediated by histone acetyltransferases and histone deacetylases enzymes that add and remove an acetyl group at the N-terminal of lysine residues, respectively. The dysregulation of these epigenetic processes could bring to the modulation of genes involved in tumorigenesis[55-57].
Non-coding RNAs: biogenesis and mechanism of action
Non-coding RNAs represent almost the totality of human genome. Only 2% of the transcribed DNA is translated into proteins, while the remnant has any protein-coding potential due to the lack of open reading frames, and it includes the transcripts so-called non-coding RNAs[58]. Based on their size, non-coding RNAs are classified into micro-RNAs (miRNAs) and long non-coding RNAs (lncRNAs)[58].
MiRNAs are endogenous transcripts of about 20-22 nucleotides, and they arise from autonomous transcriptional units or depend on the expression of their host genes[59]. Upon transcription (by RNA polymerase II or III) and a first processing (by the DROSHA complex) in the nucleus, the pre-miRNA is exported into the cytoplasm where it is further processed to the mature (guide strand) miRNA of 20-22 bp with the assistance of DICER[60]. Here, the miRNA is included in the RNA-induced silencing complex (RISC complex), which negatively regulates the gene expression by usually binding specific RNAs sequence(s) in the 3’-untranslated region (UTR) of the target gene[60]. A small group of miRNAs regulates gene expression through base pairing to other mRNA sites[60]. If the miRNA completely matches to the mRNA target sequence, the hybrid is unstable and prompts the degradation of the mRNA, whereas if the miRNA only partially matches the target sequence this results in interruption of the translation[60].
LncRNAs are transcripts with several nucleotides, greater than 200 base pairs, generally with 1000-10000 residues in length[61]. LncRNAs can be classified as (1) sense lncRNAs, which take origin from the exons of protein-coding genes; (2) antisense lncRNAs, showing complementarity to transcripts on the opposite strand; (3) intronic lncRNAs, arising from intronic region; (4) intergenic lncRNAs, sited between protein-coding genes; and (5) bidirectional lncRNAs, that share the promoter region of coding genes but they are transcribed in the opposite way[62]. Interestingly, some lncRNA are copies of coding genes that have lost the coding function because of mutations and are referred as pseudogenes[63].
Due to their length leading to base pairing, they are facilitated to assembly with DNA, RNA, and proteins, creating high-order macromolecular architectures that act as three-dimensional regulatory complexes[61]. LncRNAs may act both within the nucleus and the cytoplasm and regulate gene expression at both transcriptional and post-transcriptional level through different mechanisms[62]. In the nucleus, lncRNAs can (1) act as scaffold platforms recruiting to the DNA coding region the regulatory proteins complex to modulate positively or negatively gene expression; or (2) affect the conformation changes of chromosomes and the spliceosome activity; or (3) act as decoy, repressing gene expression by preventing the interaction between transcription factors and the promoter; or (4) inhibit the transcription by modifying allosterically RNA-binding proteins[62,64]. In the cytoplasm, lncRNAs can affect mRNA stability or its translation by acting as a sponge of microRNAs[65].
A novel group of ncRNAs, known as circular RNAs (circRNAs), has been described. CircRNAs are endogenous single-strand continuous loop structure, made of about 100 nucleotides, that show the 3’ and 5’ ends covalently linked together[66]. Thanks to the circular conformation that lacks exposed ends, circRNAs are highly stable structure very less sensitive to the nuclease degradation[67]. Although their mechanism of action is still to be clarified, circRNAs may regulate gene expression acting as cytoplasmic microRNA sponges or by sequestering RNA-binding proteins or by interfering with DNA transcription[68].
Role and regulation of autophagy in cancers affecting women
Primitive mutations and epigenetic modulation of oncogenes and of tumor suppressor genes involved in the autophagy pathway may affect the regulation and the effective role of autophagy during cancer development and progression. On one side, the autophagic process protects from cellular damage and genomic instability thus preventing neoplastic transformation[69]. In this respect, autophagy exerts its tumor-suppressive role through the elimination of reactive oxygen species, which may cause DNA damages, and through the degradation of oncogenic and toxic unfolded proteins[69]. On the contrary, autophagy may protect cancer cells from DNA and cellular damage induced by therapeutic agents (e.g., radio- and chemo-therapy) and provide pro-survival resistance to harsh conditions, such as hypoxia, nutrient starvation, and lack of growth factors[70,71]. In particular, this behavior is typical of cancer cells located in the hypoxic niche (i.e., the inner part of the tumor mass distant from blood vessels). Thus, in the late stage of cancer progression, autophagy may turn into a “tumor promoting” process by increasing chemoresistance, by ensuring the maintenance of cancer stemness and by driving tumor cells in a dormant state, which could eventually give rise to relapses and metastasis in distant organs[72]. Cancer progression reflects the dynamic changes in the permissive tumor microenvironment, in its cellular and molecular composition, as well as in the heterotypic interactions between tumor and stromal cells. Recent studies support the view that a “metabolic symbiosis” exists between cancer associated fibroblasts (CAFs) and cancer cells. Such metabolic crosstalk between stromal and cancer cells reciprocally affects autophagy regulation, which then reflects in the malignant progression of the cancer with onset of chemoresistance, relapse, and metastasis[9,73-76]. CAFs can affect autophagy in cancer cells through the release of inflammatory cytokines (e.g., IL-6 and IL-8) and oncometabolites (e.g., glutamine, lactic acid, and ketone bodies)[9,77]. It has been reported that CAFs could support the metastatic behavior of ovarian cancer, causing poor prognosis, through releasing the chemokine CXCL14 that induced an upregulation of the lncRNA LINC00092 in ovarian cancer cells where it determined a glycolytic shift[78]. Glycolytic shift has been reported to induce autophagy-dependent chemoresistance to cis-platinum therapy in ovarian cancer[79].
NcRNAs play a role in cancer development, acting as epigenetic modulator of the expression of oncogenes or tumor suppressor genes[80,81], and it is conceivable that ncRNAs released in the tumor microenvironment also contribute to the dysregulation of autophagy in cancer cells.
Both miRNAs and lncRNAs can be found in the tumor microenvironment[80,82]. CAFs can transfer their ncRNAs to the tumor microenvironment via exosome release[83-85]. In the context of cancers affecting women, it is reported that three miRNAs, namely miR-21, miR-378e, and miR-143, were increased in exosomes from CAFs as compared to those from normal fibroblasts, and these miRNAs when transferred to breast cancer cells increased their aggressiveness[85].
The clinical implication of dysregulated autophagy in cancers affecting women has been reported in several studies, both in vitro and in vivo, as well as in tumor-bearing patients. For instance, in breast cancer it has been shown that autophagy inhibition enhances tumor sensitivity to therapeutic agents[5-7]. Accordingly, in paclitaxel-resistant triple-negative breast cancer cells, cell death ensued upon down-regulating autophagy[86]. Accordingly, reduced level of BECLIN1 and LC3 expression correlated with better endocrine therapy response in hormone-responsive breast cancer[87]. In other studies, the sensitivity to anti-hormone- and chemo-therapies in breast cancer ameliorated following autophagy inhibition with chloroquine and 3-methyladenine treatments, respectively[88,89]. Similarly, chloroquine inhibition of autophagy degradation could avoid carboplatin resistance and ovarian tumor recurrence[90]. At variance, it has been shown that in ovarian cancer the low expression of BECLIN1 was a negative prognostic factor to platinum-based chemotherapy response[91], whereas the up-regulation of BNIP3, a positive regulator of mitophagy, potentiated cisplatin-sensitivity[92].
Down-regulation of autophagy has been reported in HPV-positive cervical cancers following EGFR-mediated activation of the AKT/mTOR pathway[93]. Additionally, the HPV oncoproteins E5, E6, and E7 have been shown to interfere with the autophagic machinery at different steps, either by reducing the expression of key regulators and ATG genes or by impairing the autophagosome-lysosome fusion[10]. These studies highlight the dual role of autophagy dysregulation in cancer development and in the therapeutic outcome, which might be explained by different genetic and epigenetic alterations in cancer cells that differently impact on the response to tumor microenvironment stimuli.
Non-coding RNAs regulation of autophagy in cancers affecting women
As outlined above, the role of autophagy in cancer progression is strictly dependent on genetic and epigenetic events that impinge on the integrity of the autophagic machinery and of its regulatory pathways [Figure 7].
Figure 7. Dual role of autophagy in cancer progression. Involvement of epigenetic mechanism on autophagy affects the bidirectional impact of autophagy process on cancer growth and, thus, onto the onset of metastasis, leading to cancer relapse.  : impact on;→: activation; ⟂: inhibition
: impact on;→: activation; ⟂: inhibition
During cancer evolution, these events become more frequent, adding to the clonal heterogeneity that characterizes the progression phase. Changes in the tumor microenvironment and the therapeutic treatments further increase the occurrence of genetic lesions as well as of epigenetic modulation of the genes that shape cancer cell phenotype.
Here, we schematically summarize the current knowledge on the epigenetic modulation of autophagy in female malignant tumors, with a focus on non-coding RNAs acting either on ATGs and on their regulators. The miRNAs and lncRNAs and their molecular targets involved in the autophagy pathway are reported in Tables 1 and 2, respectively. An ample set of miRNAs with either oncogenic or tumor suppressive function have been shown to affect the regulation of autophagy in cancers affecting women (reviewed in[94,95]).
The table shows the autophagy targets of miRNAs in female malignant tumors
| ATG gene | Role | miRNA | Cancer | Ref. |
|---|---|---|---|---|
| ATG2 | Phagophore formation | miR-30d | Ovarian,
Breast | [95] |
| ATG4 | LC3 maturation | miR-376b | Breast | [104] |
| let-7a | Ovarian | [94] | ||
| miR-101 | Breast | [135] | ||
| miR-34C | Breast | [98] | ||
| ATG5 | Autophagosome formation | miR-30d | Ovarian,
Breast | [95] |
| miR-181a | Breast | [136] | ||
| ATG7 | LC3 maturation | miR-204 | Ovarian | [137] |
| ATG9A | Phagophore formation | miR-29b | Ovarian | [96] |
| miR-34a | Ovarian | [94] | ||
| let-7a | Ovarian | [94] | ||
| miR-15b | Ovarian | [94] | ||
| ATG12 | Autophagosome formation | miR-30d | Ovarian,
Breast | [95] |
| ATG13 | Autophagy initiation complex | miR-15b | Ovarian | [94] |
| ATG14 | Autophagy nucleation complex | miR-152 | Ovarian | [138] |
| miR-15b | Ovarian | [94] | ||
| ATG16L | Autophagosome formation | miR-20a | Breast | [105] |
| let-7a | Ovarian | [94] | ||
| BECLIN1 | Autophagy interactome - Nucleation Complex | miR-20a | Breast | [105] |
| miR-124-3p | Breast | [139] | ||
| miR-30d | Ovarian,
Breast | [95] | ||
| miR-30a | Breast | [103] | ||
| miR-376b | Breast | [104] | ||
| BCL2 | BECLIN 1 interactor by BH3 domains | miR-7-5p | Cervical | [99] |
| BNIP3L | Mitophagy | miR-30d | Ovarian,
Breast | [95] |
| DIRAS3 (ARH-I) | BECLIN 1 interactor - Positive activator of autophagy interactome | miR-1305
miR-1260a miR-141-3p miR-424-5p miR-15a-5p miR-7-5p | Ovarian | [109] |
| DRAM1 | DNA Damage Regulated Autophagy Modulator 1; positive regulator of autophagy | miR-26b | Breast | [140] |
| LC3 | Autophagosome maturation | miR-204 | Ovarian | [137] |
| mTOR | mammalian Target of Rapamycin; negative regulator of autophagy | miR-18a | Breast | [141] |
| PTEN | Phosphatase and tensin homologue; Positive regulator of autophagy | miR-21 | Breast,
Ovarian, Cervical | [116]
[117] [118] |
| miR-17-5p | Ovarian | [142] | ||
| miR-222 | Breast | [143,144] | ||
| miR-29a | Breast | [143] | ||
| miR-205 | Endometrial | [145] | ||
| miR-19 | Breast | [146] | ||
| RAPTOR | Component of mTOR Complex 1 | miR-155 | Ovarian | [94] |
| TSC1 | Tuberous sclerosis 1; negative regulator of mTOR | miR-130a | Ovarian | [147] |
| let-7a | Ovarian | [94] | ||
| miR-15b | Ovarian | [94] | ||
| miR-21 | Ovarian | [94] | ||
| ULK1 | Autophagy initiation complex | miR-489 | Breast | [148] |
| miR-25 | Breast | [149] | ||
| UVRAG | Nucleation complex | miR-125b | Ovarian | [94] |
The table shows the autophagy targets of lncRNAs in female malignant tumors
| ATG gene/Target | Role | lnc RNA | Role | Mechanism | Cancer | Ref. |
|---|---|---|---|---|---|---|
| AMPK | AMP-activated protein kinase; positive regulator of autophagy | NBR2 | Tumor suppressor | Under stress condition, NBR2 interacts with AMPK, promoting its activation and reducing tumor development | Breast | [122] |
| ATG3 | LC3 maturation | MEG3 | Tumor suppressor | MEG3 suppresses tumorigenesis and induces autophagy by upregulating ATG3 | Ovarian | [129] |
| ATG7 | LC3 maturation | HULC | Tumor promoting | HULC overexpression promotes tumor progression and represses autophagy by inhibiting ATG7 expression | Ovarian | [134] |
| HOTAIR | Tumor promoting | HOTAIR increases cisplatin-induced autophagy via upregulating ATG7 expression | Ovarian | [132] | ||
| lncRNARP11-381N20.2 | Tumor suppressor | LncRNA inhibits autophagy induced by chemotherapy treatment | Cervical | [120] | ||
| BECLIN1 | Nucleation complex | H19 | Tumor promoting | H19 enhances tamoxifen resistance by preventing BECLIN1 methylation, thus promoting autophagy | Breast | [125] |
| HOTAIR | Tumor promoting | HOTAIR reduces cisplatin-sensitivity by inducing autophagy | Endometrial | [133] | ||
| Tumor promoting | HOTAIR promotes autophagy through Wnt pathway activation | Cervical | [121] | |||
| LC3 | Autophagosome maturation | HOTAIR | Tumor promoting | HOTAIR promotes autophagy through Wnt pathway activation | Cervical | [121] |
| lncRNARP11-381N20.2 | Tumor suppressor | LncRNA inhibits autophagy induced by chemotherapy treatment | Cervical | [120] | ||
| ULK1 | Initiation complex | GAS5 | Tumor suppressor | GAS5 inhibits cancer cell proliferation, invasion, and tumor progression by upregulating autophagy via ULK1 | Breast | [123] |
For instance, low expression of miR-29b and, consequently, high expression of its target ATG9, has been associated with ovarian cancer relapse[96]. In ovarian cancer, miR-34 is expressed at low levels, while its over-expression suppressed the NOTCH signaling pathway resulting in upregulation of autophagy and induction of apoptosis thus limiting cancer aggressiveness[97]. At variance, in cervical cancer, high levels of miR-34C-5p down-regulated autophagy, thus improving the chemotherapy response, by targeting ATG4B[98]. In cervical cancer, miR-7-5p promotes autophagy by targeting BCL-2 thus freeing BECLIN-1, and this results in chemoresistance to cisplatin[99].
The most relevant miRNAs that modulate autophagy are shown in Table 1, and include miR-30d, miR30a, miR-376b, miR-21, and miR-20a among others. Low expression of BECLIN 1, a well-known haplo-insufficient tumor suppressor, is associated with increased women’s cancer development and reduced chemotherapy response[100-102]. Both in ovarian and breast cancers, BECLIN 1 has been shown to be post-transcriptionally downregulated by several miRNAs, including miR-30a[103], and miR30d[95], miR-376b[104], and miR-20a[105]. MiR-30d is an oncomiRNA known to regulate several cellular processes, including apoptosis, senescence, proliferation, and differentiation[106]. This oncomiRNA has been shown to suppress autophagy in human ovarian and breast cancer cells by negatively affecting the expression of ATG2, ATG5, ATG12, and BNIP3, besides BECLIN 1[95]. DIRAS3 (also known as ARH-I), a well-known tumor suppressor genetically imprinted in ovarian cancer, is a positive interactor of BECLIN 1 involved in autophagy-dependent induction of cancer cell dormancy[107] and inhibition of cancer cell migration[108]. In ovarian cancer cells, DIRAS3 expression is epigenetically repressed by several predicted miRNAs, such as miR-1305, miR-1260a, miR-141-3p, miR-424-5p, miR-15a-5p, and miR-7-5p[109]. Interestingly, in ovarian cancer cells the pro-inflammatory cytokine IL-6 induces the expression of these miRNAs resulting in inhibition of DIRAS3-BECLIN 1-dependent autophagy and consequent stimulation of cell migration[109]. Besides targeting BECLIN 1, miR20a also targets ATG16 in breast cancer[107], while miR376b also targets ATG4 in breast cancer[105]. Let-7a is a tumor suppressor miRNA known to down-regulate the RAS oncogenic pathway in a variety of cancers, and its high expression correlates with chemo-responsiveness, low invasiveness, and better survival in ovarian cancer patients[110,111]. In ovarian cancer, this miRNA was shown to prevent the formation of autophagosomes by targeting ATG4 (the LC3 processing enzyme), ATG9A, and ATG16L[94]. Interestingly, autophagy degradation of p62/SQSTM1 led to decreased levels of the miRNA-processing enzyme DICER1 resulting in increased miR-Let-7a and suppression of ovarian cancer motility[112].
The dual lipid-protein phosphatase PTEN is a well-known tumor suppressor, and its deletion or silencing concurs to maintain the PI3KC1-AKT-mTORC1 pathway active, which results in down-regulation of autophagy[113-115]. In breast, ovarian, and cervical cancer, PTEN was found post-transcriptionally silenced by the oncogenic miR-21, whose high expression correlated with tumorigenesis, metastasis, and poor clinical outcome[116-118]. In ovarian cancer, both Let-7a and miR-21 were shown to target TSC1[94], an inhibitor of mTORC1.
LncRNAs, another component of ncRNA epigenetic mechanism, contribute to the development and progression of cancers acting either as tumor promoter or tumor suppressor, and many of these have been shown to affect the regulation of autophagy[119]. For instance, MEG3, PVT1, BANCR, and HNF1A-AS1 are among the inducers, while PTENP1 (the PTEN pseudogene), PCA3, and POU3F3 are among the inhibitors, while ROR and GAS5 have been reported to either induce or inhibit autophagy depending on the genetic background of the cancer cells, a fact that outlines the extremely complex network of signaling in which the ncRNAs operate[119]. Here, we report the unique lncRNAs known to modulate autophagy and that have been found to play a role in cancers affecting women. In cervical cancer, the treatment with paclitaxel upregulated the lncRNA RP11-381N20.2 that suppressed the expression of the autophagy proteins LC3 and ATG7[120]. HOTAIR downregulation, a lncRNA targeting LC3 and BECLIN1, inhibits autophagy and EMT in cervical cancer[121].
In breast cancer studies, the lncRNA NBR2 acted as a tumor suppressor since its reduction contributed to tumorigenesis and correlated with poor survival[122]. NBR2 was found to increase the expression of AMPK, an energy stress kinase sensor that enhances autophagy under nutrient deprivation condition. The lncRNA GAS5 positively regulates ULK1 expression, crucial for initiation complex formation, thus promotes autophagy in breast cancer cell lines[123]. The lncRNA H19 is known to act as an oncogene in all the three steps of carcinogenesis[124]. In breast cancer, overexpression of H19 was found to prevent BECLIN1 methylation and this correlated with autophagy-mediated resistance to hormone-therapy[125].
In ovarian cancer, several lncRNAs epigenetically control autophagy at the level of LC3 maturation. Low expression of MEG3 is associated with several cancers affecting women[126,127], suggesting that this lncRNA acts as a tumor suppressor[128]. In ovarian cancer, the ectopic overexpression of MEG3 induces autophagy through the interaction and stabilization of ATG3 mRNA and protein, and this was found to limit ovarian cancer cell proliferation[129]. On the contrary, the lncRNAs HOTAIR and HULC play a tumor promoter role. In fact, HOTAIR expression level is higher in serous ovarian and endometrial cancer tissues compared to normal tissues, making this lncRNA a biomarker associated with poor prognosis[130,131]. HOTAIR positively correlates with ATG7 expression levels, promoting autophagy-mediated chemoresistance[132]. Consistent with this finding, in endometrial cancer HOTAIR promoted BECLIN1 expression and up-regulation of autophagy, and this reduced sensitivity to chemotherapy[133]. The lncRNA HULC promotes tumorigenesis and is reportedly overexpressed in ovarian cancer, where it was found to inhibit the autophagy machinery by interacting with ATG7 and thus preventing LC3 maturation[134]. These findings again outline the double-face role of autophagy in cancer, which is tumor suppressive in the developmental stage, yet it could be tumor promoting in conferring chemoresistance in the advanced stage.
Conclusion and translational perspectives
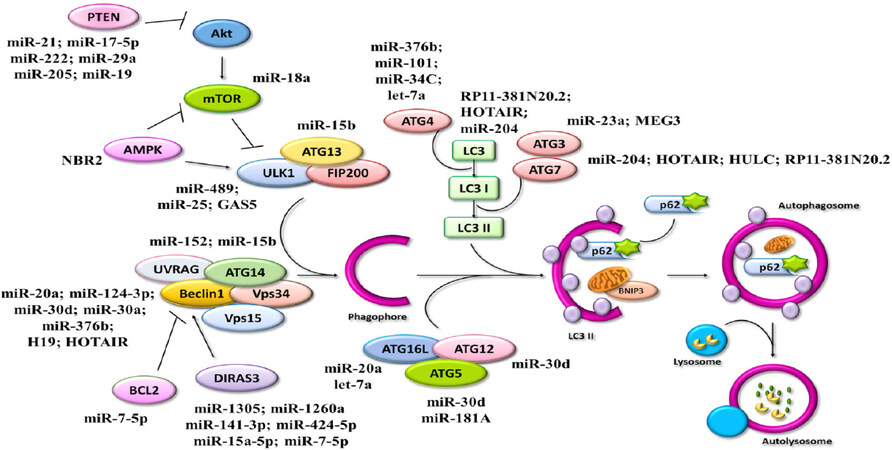
It is well-known the dual role of autophagy in cancer development. On one side, autophagy guarantees cellular homeostasis and governs cell proliferation, metabolism, invasiveness, and cell death, thus preventing the onset of malignancies. On the other hand, as pro-survival mechanism, in advanced stages autophagy supports the survival of cancer cells under harsh conditions (for instance, by promoting a dormant state) and resistance to cytotoxic therapeutic treatments. Epigenetics plays a pivotal role in controlling autophagy through the modulation of ATG genes and of their regulators, thus impacting on cancer development and progression (reviewed in[150]). This review provides a comprehensive overview of miRNAs and lncRNAs involved in the development of tumors affecting women through the induction or inhibition of autophagy process (summarized in Figure 8).
Figure 8. Cartoon showing ncRNAs and related autophagy targets in cancers affecting women. Schematic representation of miRNAs and lncRNAs and their involvement in the regulation of the autophagy pathway. →: activation; ⟂: inhibition; ncRNAs: non-coding RNAs; miRNAs: micro-RNAs; lncRNAs; long non-coding RNAs; ATGs: autophagy-related genes; PTEN: phosphatase and tensin homolog; UVRAG: UV resistance-associated gene
The question raises on how the present knowledge can be translated into the clinics. There are studies reporting on the diagnostic/prognostic potential of some ncRNAs targeting autophagy. As an example, the plasma level of miR-16, miR-27a, miR-107, miR-130a, miR-132, and miR-146a was proposed as a signature of invasive breast cancer[151]. A set of miRNAs modulating autophagy at different steps has been shown the potential as biomarker and therapeutic target in gastrointestinal cancers[152]. Similarly, the expression of the autophagy-related lncRNAs LUCAT1, AC099850.3, ZFPM2-AS1, and AC009005.1 has been proposed as a signature for the prognosis of hepatocellular carcinoma[153]. Additionally, the lncRNAs HULC (which triggers autophagy in hepatocellular carcinoma) and AC023115.3 (that inhibits autophagy in glioblastoma) have been proposed as potential biomarkers and therapeutic targets in the respective cancers[154,155]. Also, circ-ncRNAs might have diagnostic, prognostic, and therapeutic potential, though at present their mechanism of action is still obscure[156,157].
However, the data do not consistently prove the net function of the given ncRNAs in promoting or inhibiting autophagy and cancer, indicating that its action is tumor-context dependent. For instance, hyper-expression of MEG3 associated with cisplatin chemoresistance in advanced breast cancer and correlated with poor prognosis[158]. Yet, this same lncRNA acted as a tumor suppressor in ovarian cancer by upregulating ATG3 and so inducing autophagy[129].
Taken together, these data point to the need of a thorough assessment of the genetic and epigenetic signature of the cancer for designing a personalized therapy in the treatment of malignancies affecting women.
In conclusion, research on the potential exploitation of ncRNAs as therapeutic targets is still at an embryonal stage, essentially because of the complexity of the network of signaling in which they operate that makes difficult to define clearly their tumor promoting or tumor suppressive role. When referring to the impact of ncRNAs on autophagy regulation for therapeutic purposes, another layer of complexity is given by the double and opposite role of autophagy in cancer development, which again outlines the importance of the genetic and epigenetic background of the cell as well as of the signals provided by tumor microenvironment.
Thus, more studies are needed to dissect how the complex network of ncRNAs impacts on the autophagy process to identify the relevant ncRNA (and the signaling pathway) that could be used as diagnostic/prognostic biomarker or as therapeutic target. In this regard, it is worth noting that a variety of natural products (including resveratrol, curcumin, genistein, and epigallocatechin-3-gallate) have been shown to affect the expression of autophagy genes through the modulation of ncRNAs, and some of these compounds are under clinical trial evaluation as adjuvant therapeutics in a variety of cancers (reviewed in[150,159]).
Declarations
AcknowledgementsAF is recipient of a post-doctoral fellowship “Paolina Troiano” (id. 24094) granted by Associazione Italiana per la Ricerca sul Cancro (AIRC, Milan, Italy). LV and ES are PhD students recipient of a fellowship granted by the Italian Ministry of Education, University and Research (MIUR, Rome, Italy) with the contribution of Associazione per la Ricerca Medica Ippocrate-Rhazi (ARM-IR, Novara, Italy). CV was supported with a post-doctoral fellowship from Università degli Studi del Piemonte Orientale “Amedeo Avogadro” (id. 1412) granted by Fondazione Cassa di Risparmio di Torino (CRT, Torino, Italy). AS was supported with a short-term fellowship granted by Consorzio InterUniversitario per le Biotecnologie CIB (Trieste, Italy).
Authors’ contributionsMade the literature search and drafted the manuscript: Vidoni C, Vallino L, Ferraresi A, Secomandi E
Made the literature search and prepared the Tables: Salwa A, Chinthakindi M, Galetto A
Contributed to drawing the figures: Vidoni C, Vallino L
Revised and finalized the manuscript: Dhanasekaran DN, Isidoro C
Availability of data and materialsNot applicable.
Financial support and sponsorshipNone.
Conflict of interestAll authors declared that there are no conflicts of interest.
Ethical approval and consent to participateNot applicable.
Consent for publicationNot applicable.
Copyright© The Author(s) 2021.
REFERENCES
5. Han Y, Fan S, Qin T, et al. Role of autophagy in breast cancer and breast cancer stem cells (Review). Int J Oncol 2018;52:1057-70.
6. Romero MA, Bayraktar Ekmekcigil O, Bagca BG, et al. Role of autophagy in breast cancer development and progression: opposite sides of the same coin. In: Ahmad A, editor. Breast cancer metastasis and drug resistance. Cham: Springer International Publishing; 2019. pp. 65-73.
7. Lisiak N, Toton E, Rybczynska M. Autophagy as a potential therapeutic target in breast cancer treatment. Curr Cancer Drug Targets 2018;18:629-39.
8. Peracchio C, Alabiso O, Valente G, Isidoro C. Involvement of autophagy in ovarian cancer: a working hypothesis. J Ovarian Res 2012;5:22.
9. Thuwajit C, Ferraresi A, Titone R, Thuwajit P, Isidoro C. The metabolic cross-talk between epithelial cancer cells and stromal fibroblasts in ovarian cancer progression: autophagy plays a role. Med Res Rev 2018;38:1235-54.
10. Mattoscio D, Medda A, Chiocca S. Human papilloma virus and autophagy. Int J Mol Sci 2018;19:1775.
11. Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability--an evolving hallmark of cancer. Nat Rev Mol Cell Biol 2010;11:220-8.
12. Guo M, Peng Y, Gao A, Du C, Herman JG. Epigenetic heterogeneity in cancer. Biomark Res 2019;7:23.
13. Garcia-Gomez A, Rodríguez-Ubreva J, Ballestar E. Epigenetic interplay between immune, stromal and cancer cells in the tumor microenvironment. Clin Immunol 2018;196:64-71.
14. Yamashita S, Kishino T, Takahashi T, et al. Genetic and epigenetic alterations in normal tissues have differential impacts on cancer risk among tissues. Proc Natl Acad Sci U S A 2018;115:1328-33.
15. Ferlay J, Steliarova-Foucher E, Antoni S, Masuyer E. Data processing. IARC Sci Publ 2014:107-11.
16. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424.
18. Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med 2010;363:1938-48.
19. Joshi H, Press MF. Molecular oncology of breast cancer. The Breast. Elsevier; 2018. pp. 282-307.e5.
20. Tao ZQ, Shi AM, Lu CT, Song T, Zhang ZG, Zhao J. Breast Cancer: Epidemiology and Etiology. Cell Biochem Biophys 2015;72:333-8.
21. Mahdavi M, Nassiri M, Kooshyar MM, et al. Hereditary breast cancer; Genetic penetrance and current status with BRCA. J Cell Physiol 2019;234:5741-50.
22. Reid BM, Permuth JB, Sellers TA. Epidemiology of ovarian cancer: a review. Cancer Biol Med 2017;14:9-32.
23. Ren F, Shen J, Shi H, Hornicek FJ, Kan Q, Duan Z. Novel mechanisms and approaches to overcome multidrug resistance in the treatment of ovarian cancer. Biochim Biophys Acta 2016;1866:266-75.
24. Koshiyama M, Matsumura N, Konishi I. Recent concepts of ovarian carcinogenesis: type I and type II. Biomed Res Int 2014;2014:934261.
25. Toss A, Tomasello C, Razzaboni E, et al. Hereditary ovarian cancer: not only BRCA 1 and 2 genes. Biomed Res Int 2015;2015:11.
26. Stewart C, Ralyea C, Lockwood S. Ovarian cancer: an integrated review. Semin Oncol Nurs 2019;35:151-6.
27. Chandra A, Pius C, Nabeel M, et al. Ovarian cancer: current status and strategies for improving therapeutic outcomes. Cancer Med 2019;8:7018-31.
28. Casey MJ, Summers GK, Crotzer D. Endometrial Cancer. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2020. Available from: https://www.ncbi.nlm.nih.gov/books/NBK525981/. [Last accessed on 24 Dec 2020].
29. Matias-Guiu X, Prat J. Molecular pathology of endometrial carcinoma. Histopathology 2013;62:111-23.
30. Dowty JG, Win AK, Buchanan DD, et al. Cancer risks for MLH1 and MSH2 mutation carriers. Hum Mutat 2013;34:490-7.
31. Kurman RJ, Carcangiu ML, Herrington CS, Young RH. WHO classification of tumors of the female reproductive organs. WHO classification of tumors. Lyon: IARC press; 2014.
32. Santaballa A, Matías-Guiu X, Redondo A, et al. SEOM clinical guidelines for endometrial cancer (2017). Clin Transl Oncol 2018;20:29-37.
33. Passarello K, Kurian S, Villanueva V. Endometrial Cancer: An Overview of Pathophysiology, Management, and Care. Semin Oncol Nurs 2019;35:157-65.
34. Duenas-Gonzalez A, Gonzalez-Fierro A. Pharmacodynamics of current and emerging treatments for cervical cancer. Expert Opin Drug Metab Toxicol 2019;15:671-82.
35. Fowler JR, Jack BW. Cancer, Cervical. [Updated 2019 Sep 9]. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2020. Available from: https://www.ncbi.nlm.nih.gov/books/NBK431093/. [Last accessed on 24 Dec 2020].
36. Wang XL, Huang XM, Zhang YZ. Involvement of human papillomaviruses in cervical cancer. Front Microbiol 2018;9:2896.
37. Ghosh I, Mandal R, Kundu P, Biswas J. Association of genital infections other than human papillomavirus with pre-invasive and invasive cervical neoplasia. J Clin Diagn Res 2016;10:XE01-6.
39. Weidberg H, Shvets E, Elazar Z. Biogenesis and cargo selectivity of autophagosomes. Annu Rev Biochem 2011;80:125-56.
40. Galluzzi L, Baehrecke EH, Ballabio A, et al. Molecular definitions of autophagy and related processes. EMBO J 2017;36:1811-36.
42. Ravanan P, Srikumar IF, Talwar P. Autophagy: the spotlight for cellular stress responses. Life Sci 2017;188:53-67.
43. Hansen M, Rubinsztein DC, Walker DW. Autophagy as a promoter of longevity: insights from model organisms. Nat Rev Mol Cell Biol 2018;19:579-93.
44. Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett 1993;333:169-74.
45. Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med 2013;368:651-62.
46. Yu L, Chen Y, Tooze SA. Autophagy pathway: cellular and molecular mechanisms. Autophagy 2018;14:207-15.
47. Saha S, Panigrahi DP, Patil S, Bhutia SK. Autophagy in health and disease: a comprehensive review. Biomed Pharmacother 2018;104:485-95.
48. Huang T, Song X, Yang Y, et al. Autophagy and hallmarks of cancer. Crit Rev Oncog 2018;23:247-67.
49. Corona Velazquez AF, Jackson WT. So many roads: the multifaceted regulation of autophagy induction. Mol Cell Biol 2018;38:e00303-18.
50. Follo C, Vidoni C, Morani F, Ferraresi A, Seca C, Isidoro C. Amino acid response by Halofuginone in Cancer cells triggers autophagy through proteasome degradation of mTOR. Cell Commun Signal 2019;17:39.
51. Polson HE, de Lartigue J, Rigden DJ, et al. Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy 2010;6:506-22.
52. Geng J, Klionsky DJ. The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. ‘Protein modifications: beyond the usual suspects’ review series. EMBO Rep 2008;9:859-64.
53. Feinberg AP. The Key Role of epigenetics in human disease prevention and mitigation. N Engl J Med 2018;378:1323-34.
54. Long HK, King HW, Patient RK, Odom DT, Klose RJ. Protection of CpG islands from DNA methylation is DNA-encoded and evolutionarily conserved. Nucleic Acids Res 2016;44:6693-706.
55. Seto E, Yoshida M. Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harb Perspect Biol 2014;6:a018713.
56. Sanaei M, Kavoosi F. Histone Deacetylases and histone deacetylase inhibitors: molecular mechanisms of action in various cancers. Adv Biomed Res 2019;8:63.
57. Borley J, Brown R. Epigenetic mechanisms and therapeutic targets of chemotherapy resistance in epithelial ovarian cancer. Ann Med 2015;47:359-69.
58. Djebali S, Davis CA, Merkel A, et al. Landscape of transcription in human cells. Nature 2012;489:101-8.
59. Ozsolak F, Poling LL, Wang Z, et al. Chromatin structure analyses identify miRNA promoters. Genes Dev 2008;22:3172-83.
60. O’Brien J, Hayder H, Zayed Y, Peng C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol (Lausanne) 2018;9:402.
61. Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature 2012;482:339-46.
62. Dykes IM, Emanueli C. Transcriptional and post-transcriptional gene regulation by long non-coding RNA. Genomics Proteomics Bioinformatics 2017;15:177-86.
63. Milligan MJ, Lipovich L. Pseudogene-derived lncRNAs: emerging regulators of gene expression. Front Genet 2014;5:476.
64. Wei JW, Huang K, Yang C, Kang CS. Non-coding RNAs as regulators in epigenetics (Review). Oncol Rep 2017;37:3-9.
65. Noh JH, Kim KM, McClusky WG, Abdelmohsen K, Gorospe M. Cytoplasmic functions of long noncoding RNAs. Wiley Interdiscip Rev RNA 2018;9:e1471.
67. Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013;19:141-57.
68. Ebbesen KK, Kjems J, Hansen TB. Circular RNAs: identification, biogenesis and function. Biochim Biophys Acta 2016;1859:163-8.
69. Kocaturk NM, Akkoc Y, Kig C, Bayraktar O, Gozuacik D, Kutlu O. Autophagy as a molecular target for cancer treatment. Eur J Pharm Sci 2019;134:116-37.
70. Maycotte P, Jones KL, Goodall ML, Thorburn J, Thorburn A. Autophagy supports breast cancer stem cell maintenance by regulating IL6 secretion. Mol Cancer Res 2015;13:651-8.
72. Vera-Ramirez L, Vodnala SK, Nini R, Hunter KW, Green JE. Autophagy promotes the survival of dormant breast cancer cells and metastatic tumour recurrence. Nat Commun 2018;9:1944.
73. Gomes LR, Vessoni AT, Menck CFM. Microenvironment and autophagy cross-talk: Implications in cancer therapy. Pharmacol Res 2016;107:300-7.
74. Martinez-Outschoorn U, Sotgia F, Lisanti MP. Tumor microenvironment and metabolic synergy in breast cancers: critical importance of mitochondrial fuels and function. Semin Oncol 2014;41:195-216.
75. Martinez-Outschoorn UE, Lisanti MP, Sotgia F. Catabolic cancer-associated fibroblasts transfer energy and biomass to anabolic cancer cells, fueling tumor growth. Semin Cancer Biol 2014;25:47-60.
76. Ferraresi A, Girone C, Esposito A, et al. How autophagy shapes the tumor microenvironment in ovarian cancer. Front Oncol 2020;10:599915.
77. Yan YL, Chen X, Wang X, et al. The effects and the mechanisms of autophagy on the cancer-associated fibroblasts in cancer. J Exp Clin Cancer Res 2019;38:171.
78. Zhao LJ, Ji GL, Le XB, et al. Long noncoding RNA LINC00092 acts in cancer-associated fibroblasts to drive glycolysis and progression of ovarian cancer. Cancer Res 2017;77:1369-82.
79. Zhang XY, Zhang M, Cong Q, et al. Hexokinase 2 confers resistance to cisplatin in ovarian cancer cells by enhancing cisplatin-induced autophagy. Int J Biochem Cell Biol 2018;95:9-16.
80. Evans JR, Feng FY, Chinnaiyan AM. The bright side of dark matter: lncRNAs in cancer. J Clin Invest 2016;126:2775-82.
81. Mendell JT. MicroRNAs: critical regulators of development, cellular physiology and malignancy. Cell Cycle 2005;4:1179-84.
82. Patil N, Allgayer H, Leupold JH. MicroRNAs in the tumor microenvironment. In: Birbrair A, editor. Tumor Microenvironment. Cham: Springer International Publishing; 2020. pp. 1-31.
83. Ren J, Ding L, Zhang D, et al. Carcinoma-associated fibroblasts promote the stemness and chemoresistance of colorectal cancer by transferring exosomal lncRNA H19. Theranostics 2018;8:3932-48.
84. Herrera M, Llorens C, Rodríguez M, et al. Differential distribution and enrichment of non-coding RNAs in exosomes from normal and Cancer-associated fibroblasts in colorectal cancer. Mol Cancer 2018;17:114.
85. Donnarumma E, Fiore D, Nappa M, et al. Cancer-associated fibroblasts release exosomal microRNAs that dictate an aggressive phenotype in breast cancer. Oncotarget 2017;8:19592-608.
86. Wen J, Yeo S, Wang CR, et al. Autophagy inhibition re-sensitizes pulse stimulation-selected paclitaxel-resistant triple negative breast cancer cells to chemotherapy-induced apoptosis. Breast Cancer Res Treat 2015;149:619-29.
87. Ueno T, Saji S, Sugimoto M, et al. Clinical significance of the expression of autophagy-associated marker, beclin 1, in breast cancer patients who received neoadjuvant endocrine therapy. BMC Cancer 2016;16:230.
88. Cook KL, Wärri A, Soto-Pantoja DR, et al. Hydroxychloroquine inhibits autophagy to potentiate antiestrogen responsiveness in ER+ breast cancer. Clin Cancer Res 2014;20:3222-32.
89. Aydinlik S, Erkisa M, Cevatemre B, et al. Enhanced cytotoxic activity of doxorubicin through the inhibition of autophagy in triple negative breast cancer cell line. Biochim Biophys Acta Gen Subj 2017;1861:49-57.
90. Pagotto A, Pilotto G, Mazzoldi EL, et al. Autophagy inhibition reduces chemoresistance and tumorigenic potential of human ovarian cancer stem cells. Cell Death Dis 2017;8:e2943.
91. Minamoto T, Nakayama K, Nakamura K, et al. Loss of beclin 1 expression in ovarian cancer: A potential biomarker for predicting unfavorable outcomes. Oncol Lett 2018;15:1170-6.
92. Jia JH, Yang XX, Zhao Q, et al. BNIP3 contributes to cisplatin-induced apoptosis in ovarian cancer cells. FEBS Open Bio 2020;10:1463-73.
93. Surviladze Z, Sterk RT, DeHaro SA, Ozbun MA. Cellular entry of human papillomavirus type 16 involves activation of the phosphatidylinositol 3-kinase/Akt/mTOR pathway and inhibition of autophagy. J Virol 2013;87:2508-17.
94. Titone R, Morani F, Follo C, Vidoni C, Mezzanzanica D, Isidoro C. Epigenetic control of autophagy by microRNAs in ovarian cancer. Biomed Res Int 2014;2014:343542.
95. Yang XJ, Zhong XM, Tanyi JL, et al. MiR-30d Regulates multiple genes in the autophagy pathway and impairs autophagy process in human cancer cells. Biochem Biophys Res Commun 2013;431:617-22.
96. Dai FR, Zhang Y, Chen YX. Involvement of miR-29b signaling in the sensitivity to chemotherapy in patients with ovarian carcinoma. Hum Pathol 2014;45:1285-93.
97. Jia Y, Lin RX, Jin HJ, et al. MicroRNA-34 suppresses proliferation of human ovarian cancer cells by triggering autophagy and apoptosis and inhibits cell invasion by targeting Notch 1. Biochimie 2019;160:193-9.
98. Wu YR, Ni ZH, Yan XJ, et al. Targeting the MIR34C-5p-ATG4B-autophagy axis enhances the sensitivity of cervical cancer cells to pirarubicin. Autophagy 2016;12:1105-17.
99. Yang FX, Guo L, Cao YJ, Li SZ, Li JH, Liu M. MicroRNA-7-5p promotes cisplatin resistance of cervical cancer cells and modulation of cellular energy homeostasis by regulating the expression of the PARP-1 and BCL2 genes. Med Sci Monit 2018;24:6506-16.
100. Tang H, Sebti S, Titone R, et al. Decreased BECN1 mRNA Expression in human breast cancer is associated with estrogen receptor-negative subtypes and poor prognosis. EBioMedicine 2015;2:255-63.
101. Lin HX, Qiu HJ, Zeng F, et al. Decreased expression of Beclin 1 correlates closely with Bcl-xL expression and poor prognosis of ovarian carcinoma. PLoS One 2013;8:e60516.
102. Valente G, Morani F, Nicotra G, et al. Expression and clinical significance of the autophagy proteins BECLIN 1 and LC3 in ovarian cancer. Biomed Res Int 2014;2014:462658.
103. Zhu H, Wu H, Liu XP, et al. Regulation of autophagy by a beclin 1-targeted microRNA, miR-30a, in cancer cells. Autophagy 2009;5:816-23.
104. Korkmaz G, le Sage C, Tekirdag KA, Agami R, Gozuacik D. miR-376b controls starvation and mTOR inhibition-related autophagy by targeting ATG4C and BECN1. Autophagy 2012;8:16576.
105. Liu L, He J, Wei X, et al. MicroRNA-20a-mediated loss of autophagy contributes to breast tumorigenesis by promoting genomic damage and instability. Oncogene 2017;36:5874-84.
106. Mao L, Liu SM, Hu L, et al. MiR-30 family: a promising regulator in development and disease. Biomed Res Int 2018;2018:9623412.
107. Lu Z, Baquero MT, Yang H, et al. DIRAS3 regulates the autophagosome initiation complex in dormant ovarian cancer cells. Autophagy 2014;10:1071-92.
108. Lu Z, Bast RC Jr. The tumor suppressor gene ARHI (DIRAS3) inhibits ovarian cancer cell migration through multiple mechanisms. Cell Adh Migr 2013;7:232-6.
109. Ferraresi A, Phadngam S, Morani F, et al. Resveratrol inhibits IL-6-induced ovarian cancer cell migration through epigenetic up-regulation of autophagy. Mol Carcinog 2017;56:1164-81.
110. Qiu L, Zhang GF, Yu L, Wang HY, Jia XJ, Wang TJ. Novel oncogenic and chemoresistance-inducing functions of resistin in ovarian cancer cells require miRNAs-mediated induction of epithelial-to-mesenchymal transition. Sci Rep 2018;8:12522.
111. Lu L, Schwartz P, Scarampi L, et al. MicroRNA let-7a: a potential marker for selection of paclitaxel in ovarian cancer management. Gynecol Oncol 2011;122:366-71.
112. Liao CC, Ho MY, Liang SM, Liang CM. Autophagic degradation of SQSTM1 inhibits ovarian cancer motility by decreasing DICER1 and AGO2 to induce MIRLET7A-3P. Autophagy 2018;14:2065-82.
113. Arico S, Petiot A, Bauvy C, et al. The tumor suppressor PTEN positively regulates macroautophagy by inhibiting the phosphatidylinositol 3-kinase/protein kinase B pathway. J Biol Chem 2001;276:35243-46.
114. Morani F, Phadngam S, Follo C, et al. PTEN deficiency and mutant p53 confer glucose-addiction to thyroid cancer cells: impact of glucose depletion on cell proliferation, cell survival, autophagy and cell migration. Genes Cancer 2014;5:226-39.
115. Phadngam S, Castiglioni A, Ferraresi A, Morani F, Follo C, Isidoro C. PTEN dephosphorylates AKT to prevent the expression of GLUT1 on plasmamembrane and to limit glucose consumption in cancer cells. Oncotarget 2016;7:84999-5020.
116. Yu XF, Li RL, Shi WN, et al. Silencing of MicroRNA-21 confers the sensitivity to tamoxifen and fulvestrant by enhancing autophagic cell death through inhibition of the PI3K-AKT-mTOR pathway in breast cancer cells. Biomed Pharmacother 2016;77:37-44.
117. Lou YH, Yang XS, Wang FL, Cui ZM, Huang Y. MicroRNA-21 promotes the cell proliferation, invasion and migration abilities in ovarian epithelial carcinomas through inhibiting the expression of PTEN protein. Int J Mol Med 2010;26:819-27.
118. Peralta-Zaragoza O, Deas J, Meneses-Acosta A, et al. Relevance of miR-21 in regulation of tumor suppressor gene PTEN in human cervical cancer cells. BMC Cancer 2016;16:215.
119. Islam Khan MZ, Tam SY, Law HKW. Autophagy-modulating long non-coding RNAs (LncRNAs) and their molecular rvents in cancer. Front Genet 2018;9:750.
120. Zou SH, Du X, Lin H, Wang PC, Li M. Paclitaxel inhibits the progression of cervical cancer by inhibiting autophagy via lncRNARP11-381N20.2. Eur Rev Med Pharmacol Sci 2018;22:3010-7.
121. Guo XG, Xiao HQ, Guo SH, et al. Long noncoding RNA HOTAIR knockdown inhibits autophagy and epithelial-mesenchymal transition through the Wnt signaling pathway in radioresistant human cervical cancer HeLa cells. J Cell Physiol 2019;234:3478-89.
122. Liu XW, Xiao ZD, Han L, et al. LncRNA NBR2 engages a metabolic checkpoint by regulating AMPK under energy stress. Nat Cell Biol 2016;18:431-42.
123. Li GP, Qian L, Tang XQ, Chen Y, Zhao Z, Zhang CW. Long non-coding RNA growth arrest-specific 5 (GAS5) acts as a tumor suppressor by promoting autophagy in breast cancer. Mol Med Rep 2020;22:2460-8.
124. Raveh E, Matouk IJ, Gilon M, Hochberg A. The H19 Long non-coding RNA in cancer initiation, progression and metastasis - a proposed unifying theory. Mol Cancer 2015;14:184.
125. Wang J, Xie SD, Yang JJ, et al. The long noncoding RNA H19 promotes tamoxifen resistance in breast cancer via autophagy. J Hematol Oncol 2019;12:81.
126. Sun L, Li Y, Yang BX. Downregulated long non-coding RNA MEG3 in breast cancer regulates proliferation, migration and invasion by depending on p53’s transcriptional activity. Biochem Biophys Res Commun 2016;478:323-9.
127. Qin R, Chen Z, Ding Y, Hao J, Hu J, Guo F. Long non-coding RNA MEG3 inhibits the proliferation of cervical carcinoma cells through the induction of cell cycle arrest and apoptosis. Neoplasma 2013;60:486-92.
128. Zhang X, Zhou YL, Mehta KR, et al. A pituitary-derived MEG3 isoform functions as a growth suppressor in tumor cells. J Clin Endocrinol Metab 2003;88:5119-26.
129. Xiu YL, Sun KX, Chen X, et al. Upregulation of the lncRNA Meg3 induces autophagy to inhibit tumorigenesis and progression of epithelial ovarian carcinoma by regulating activity of ATG3. Oncotarget 2017;8:31714-25.
130. Qiu JJ, Wang Y, Ding JX, Jin HY, Yang G, Hua KQ. The long non-coding RNA HOTAIR promotes the proliferation of serous ovarian cancer cells through the regulation of cell cycle arrest and apoptosis. Exp Cell Res 2015;333:238-48.
131. He XY, Bao W, Li XC, et al. The long non-coding RNA HOTAIR is upregulated in endometrial carcinoma and correlates with poor prognosis. Int J Mol Med 2014;33:325-32.
132. Yu Y, Zhang XH, Tian HS, Zhang ZY, Tian YJ. Knockdown of long non-coding RNA HOTAIR increases cisplatin sensitivity in ovarian cancer by inhibiting cisplatin-induced autophagy. J BUON 2018;23:1396-401.
133. Sun MY, Zhu JY, Zhang CY, et al. Autophagy regulated by lncRNA HOTAIR contributes to the cisplatin-induced resistance in endometrial cancer cells. Biotechnol Lett 2017;39:1477-84.
134. Chen S, Wu DD, Sang XB, et al. The lncRNA HULC functions as an oncogene by targeting ATG7 and ITGB1 in epithelial ovarian carcinoma. Cell Death Dis 2017;8:e3118.
135. Frankel LB, Wen J, Lees M, et al. MicroRNA-101 is a potent inhibitor of autophagy. EMBO J 2011;30:4628-41.
136. Tekirdag KA, Korkmaz G, Ozturk DG, Agami R, Gozuacik D. MiR-181A regulates starvation- and rapamycin-induced autophagy through targeting of ATG5. Autophagy 2013;9:374-85.
137. Tang J, Zhu J, Ye YG, et al. Inhibition LC3B can increase chemosensitivity of ovarian cancer cells. Cancer Cell Int 2019;19:199.
138. He J, Yu JJ, Xu Q, et al. Downregulation of ATG14 by EGR1-MIR152 sensitizes ovarian cancer cells to cisplatin-induced apoptosis by inhibiting cyto-protective autophagy. Autophagy 2015;11:373-84.
139. Zhang FF, Wang BB, Long HL, et al. Decreased miR-124-3p expression prompted breast cancer cell progression mainly by targeting beclin-1. Clin Lab 2016;62:1139-45.
140. Meng CD, Liu Y, Shen YN, et al. MicroRNA-26b suppresses autophagy in breast cancer cells by targeting DRAM1 mRNA, and is downregulated by irradiation. Oncol Lett 2018;15:1435-40.
141. Fan YX, Dai YZ, Wang XL, et al. MiR-18a upregulation enhances autophagy in triple negative cancer cells via inhibiting mTOR signaling pathway. Eur Rev Med Pharmacol Sci 2016;20:2194-200.
142. Fang Y, Xu CY, Fu Y. MicroRNA-17-5p induces drug resistance and invasion of ovarian carcinoma cells by targeting PTEN signaling. J Biol Res (Thessalon) 2015;22:12.
143. Zhong SL, Li WJ, Chen ZY, Xu JJ, Zhao JH. MiR-222 and miR-29a contribute to the drug-resistance of breast cancer cells. Gene 2013;531:8-14.
144. Shen HY, Wang DD, Li LP, et al. MiR-222 promotes drug-resistance of breast cancer cells to adriamycin via modulation of PTEN/Akt/FOXO1 pathway. Gene 2017;596:110-8.
145. Zhuo ZH, Yu HM. MiR-205 inhibits cell growth by targeting AKT-mTOR signaling in progesterone-resistant endometrial cancer Ishikawa cells. Oncotarget 2017;8:28042-51.
146. Wu JS, Jiang Y, Cao WS, et al. MiR-19 targeting of PTEN mediates butyl benzyl phthalate-induced proliferation in both ER(+) and ER(-) breast cancer cells. Toxicol Lett 2018;295:124-33.
147. Wang YQ, Zhang XY, Tang W, et al. MiR-130a upregulates mTOR pathway by targeting TSC1 and is transactivated by NF-κB in high-grade serous ovarian carcinoma. Cell Death Differ 2017;24:2089-100.
148. Soni M, Patel Y, Markoutsa E, et al. Autophagy, cell viability, and chemoresistance are regulated by miR-489 in breast cancer. Mol Cancer Res 2018;16:1348-60.
149. Wang ZY, Wang N, Liu PX, et al. MicroRNA-25 regulates chemoresistance-associated autophagy in breast cancer cells, a process modulated by the natural autophagy inducer isoliquiritigenin. Oncotarget 2014;5:7013-26.
150. Vidoni C, Ferraresi A, Secomandi E, Vallino L, Dhanasekaran DN, Isidoro C. Epigenetic targeting of autophagy for cancer prevention and treatment by natural compounds. Semin Cancer Biol 2020;66:34-44.
151. Stückrath I, Rack B, Janni W, Jäger B, Pantel K, Schwarzenbach H. Aberrant plasma levels of circulating miR-16, miR-107, miR-130a and miR-146a are associated with lymph node metastasis and receptor status of breast cancer patients. Oncotarget 2015;6:13387-401.
152. Pourhanifeh MH, Vosough M, Mahjoubin-Tehran M, et al. Autophagy-related microRNAs: possible regulatory roles and therapeutic potential in and gastrointestinal cancers. Pharmacol Res 2020;161:105133.
153. Wu H, Liu TT, Qi JN, Qin CY, Zhu Q. Four autophagy-related lncRNAs predict the prognosis of HCC through coexpression and ceRNA mechanism. Biomed Res Int 2020;2020:3801748.
154. Xiong H, Ni Z, He J, et al. LncRNA HULC triggers autophagy via stabilizing Sirt1 and attenuates the chemosensitivity of HCC cells. Oncogene 2017;36:3528-40.
155. Ma BB, Yuan ZB, Zhang L, et al. Long non-coding RNA AC023115.3 suppresses chemoresistance of glioblastoma by reducing autophagy. Biochim Biophys Acta Mol Cell Res 2017;1864:1393-404.
156. Qi P, Zhou XY, Du X. Circulating long non-coding RNAs in cancer: current status and future perspectives. Mol Cancer 2016;15:39.
157. Sun T. Long noncoding RNAs act as regulators of autophagy in cancer. Pharmacol Res 2018;129:151-5.
158. Bayarmaa B, Wu ZP, Peng J, et al. Association of LncRNA MEG3 polymorphisms with efficacy of neoadjuvant chemotherapy in breast cancer. BMC Cancer 2019;19:877.
Cite This Article
Export citation file: BibTeX | RIS
OAE Style
Vidoni C, Vallino L, Ferraresi A, Secomandi E, Salwa A, Chinthakindi M, Galetto A, Dhanasekaran DN, Isidoro C. Epigenetic control of autophagy in women's tumors: role of non-coding RNAs. J Cancer Metastasis Treat 2021;7:4. http://dx.doi.org/10.20517/2394-4722.2020.95
AMA Style
Vidoni C, Vallino L, Ferraresi A, Secomandi E, Salwa A, Chinthakindi M, Galetto A, Dhanasekaran DN, Isidoro C. Epigenetic control of autophagy in women's tumors: role of non-coding RNAs. Journal of Cancer Metastasis and Treatment. 2021; 7: 4. http://dx.doi.org/10.20517/2394-4722.2020.95
Chicago/Turabian Style
Vidoni, Chiara, Letizia Vallino, Alessandra Ferraresi, Eleonora Secomandi, Amreen Salwa, Menaka Chinthakindi, Alessandra Galetto, Danny N Dhanasekaran, Ciro Isidoro. 2021. "Epigenetic control of autophagy in women's tumors: role of non-coding RNAs" Journal of Cancer Metastasis and Treatment. 7: 4. http://dx.doi.org/10.20517/2394-4722.2020.95
ACS Style
Vidoni, C.; Vallino L.; Ferraresi A.; Secomandi E.; Salwa A.; Chinthakindi M.; Galetto A.; Dhanasekaran DN.; Isidoro C. Epigenetic control of autophagy in women's tumors: role of non-coding RNAs. J. Cancer. Metastasis. Treat. 2021, 7, 4. http://dx.doi.org/10.20517/2394-4722.2020.95
About This Article
Special Issue
Copyright
Data & Comments
Data
 Cite This Article 46 clicks
Cite This Article 46 clicks












Comments
Comments must be written in English. Spam, offensive content, impersonation, and private information will not be permitted. If any comment is reported and identified as inappropriate content by OAE staff, the comment will be removed without notice. If you have any queries or need any help, please contact us at support@oaepublish.com.